Defined tumor antigen-specific T cells potentiate personalized TCR-T cell therapy and prediction of immunotherapy response
- PMID: 35165422
- PMCID: PMC9160085
- DOI: 10.1038/s41422-022-00627-9
Defined tumor antigen-specific T cells potentiate personalized TCR-T cell therapy and prediction of immunotherapy response
Abstract
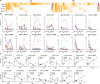
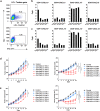
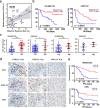
Personalized immunotherapy targeting tumor-specific antigens (TSAs) could generate efficient and safe antitumor immune response without damaging normal tissues. Although neoantigen vaccines have shown therapeutic effect in clinic trials, precise prediction of neoantigens from tumor mutations is still challenging. The host antitumor immune response selects and activates T cells recognizing tumor antigens. Hence, T cells engineered with T-cell receptors (TCRs) from these naturally occurring tumor antigen-specific T (Tas) cells in a patient will target personal TSAs in his/her tumor. To establish such a personalized TCR-T cell therapy, we comprehensively characterized T cells in tumor and its adjacent tissues by single-cell mRNA sequencing (scRNA-seq), TCR sequencing (TCR-seq) and in vitro neoantigen stimulation. Compared to bystander T cells circulating among tissues, Tas cells were characterized by tumor enrichment, tumor-specific clonal expansion and neoantigen specificity. We found that CXCL13 is a unique marker for both CD4+ and CD8+ Tas cells. Importantly, TCR-T cells expressing TCRs from Tas cells showed significant therapeutic effects on autologous patient-derived xenograft (PDX) tumors. Intratumoral Tas cell levels measured by CXCL13 expression precisely predicted the response to immune checkpoint blockade, indicating a critical role of Tas cells in the antitumor immunity. We further identified CD200 and ENTPD1 as surface markers for CD4+ and CD8+ Tas cells respectively, which enabled the isolation of Tas cells from tumor by Fluorescence Activating Cell Sorter (FACS) sorting. Overall, our results suggest that TCR-T cells engineered with Tas TCRs are a promising agent for personalized immunotherapy, and intratumoral Tas cell levels determine the response to immunotherapy.
© 2022. The Author(s) under exclusive licence to Center for Excellence in Molecular Cell Science, CAS.
Conflict of interest statement
C.B., X.L., H.L. are current employee of FineImmune. The other authors declare that they have no competing interests.
Figures



Comment in
-
Amplifying natural antitumor immunity for personalized immunotherapy.Cell Res. 2022 Jun;32(6):505-506. doi: 10.1038/s41422-022-00649-3. Cell Res. 2022. PMID: 35352030 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

