A human brain vascular atlas reveals diverse mediators of Alzheimer's risk
- PMID: 35165441
- PMCID: PMC9635042
- DOI: 10.1038/s41586-021-04369-3
A human brain vascular atlas reveals diverse mediators of Alzheimer's risk
Abstract
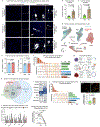
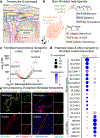
The human brain vasculature is of great medical importance: its dysfunction causes disability and death1, and the specialized structure it forms-the blood-brain barrier-impedes the treatment of nearly all brain disorders2,3. Yet so far, we have no molecular map of the human brain vasculature. Here we develop vessel isolation and nuclei extraction for sequencing (VINE-seq) to profile the major vascular and perivascular cell types of the human brain through 143,793 single-nucleus transcriptomes from 25 hippocampus and cortex samples of 9 individuals with Alzheimer's disease and 8 individuals with no cognitive impairment. We identify brain-region- and species-enriched genes and pathways. We reveal molecular principles of human arteriovenous organization, recapitulating a gradual endothelial and punctuated mural cell continuum. We discover two subtypes of human pericytes, marked by solute transport and extracellular matrix (ECM) organization; and define perivascular versus meningeal fibroblast specialization. In Alzheimer's disease, we observe selective vulnerability of ECM-maintaining pericytes and gene expression patterns that implicate dysregulated blood flow. With an expanded survey of brain cell types, we find that 30 of the top 45 genes that have been linked to Alzheimer's disease risk by genome-wide association studies (GWASs) are expressed in the human brain vasculature, and we confirm this by immunostaining. Vascular GWAS genes map to endothelial protein transport, adaptive immune and ECM pathways. Many are microglia-specific in mice, suggesting a partial evolutionary transfer of Alzheimer's disease risk. Our work uncovers the molecular basis of the human brain vasculature, which will inform our understanding of overall brain health, disease and therapy.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Figures














Comment in
-
Transcriptomic mapping of the human cerebrovasculature.Nat Rev Neurol. 2022 Jun;18(6):319-320. doi: 10.1038/s41582-022-00650-9. Nat Rev Neurol. 2022. PMID: 35322261 No abstract available.
-
The importance of genomics in advancing the diagnosis and treatment of dementia.Lancet Neurol. 2022 Aug;21(8):676-677. doi: 10.1016/S1474-4422(22)00234-4. Epub 2022 Jun 10. Lancet Neurol. 2022. PMID: 35697056 Free PMC article. No abstract available.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

