M24B aminopeptidase inhibitors selectively activate the CARD8 inflammasome
- PMID: 35165443
- PMCID: PMC9179932
- DOI: 10.1038/s41589-021-00964-7
M24B aminopeptidase inhibitors selectively activate the CARD8 inflammasome
Abstract
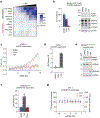
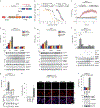
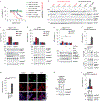
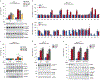
Inflammasomes are multiprotein complexes that sense intracellular danger signals and induce pyroptosis. CARD8 and NLRP1 are related inflammasomes that are repressed by the enzymatic activities and protein structures of the dipeptidyl peptidases 8 and 9 (DPP8/9). Potent DPP8/9 inhibitors such as Val-boroPro (VbP) activate both NLRP1 and CARD8, but chemical probes that selectively activate only one have not been identified. Here we report a small molecule called CQ31 that selectively activates CARD8. CQ31 inhibits the M24B aminopeptidases prolidase (PEPD) and Xaa-Pro aminopeptidase 1 (XPNPEP1), leading to the accumulation of proline-containing peptides that inhibit DPP8/9 and thereby activate CARD8. NLRP1 is distinct from CARD8 in that it directly contacts DPP8/9's active site; these proline-containing peptides, unlike VbP, do not disrupt this repressive interaction and thus do not activate NLRP1. We expect that CQ31 will now become a valuable tool to study CARD8 biology.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Figures
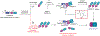



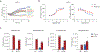



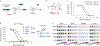


Comment in
-
XaaP-ing DPP8/9 for CARD8 activation.Nat Chem Biol. 2022 May;18(5):439-440. doi: 10.1038/s41589-021-00958-5. Nat Chem Biol. 2022. PMID: 35165444 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

