Evolutionary dynamics of SARS-CoV-2 circulating in Yogyakarta and Central Java, Indonesia: sequence analysis covering furin cleavage site (FCS) region of the spike protein
- PMID: 35165816
- PMCID: PMC8853438
- DOI: 10.1007/s10123-022-00239-8
Evolutionary dynamics of SARS-CoV-2 circulating in Yogyakarta and Central Java, Indonesia: sequence analysis covering furin cleavage site (FCS) region of the spike protein
Abstract
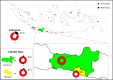
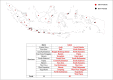
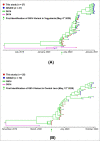
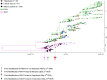
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a new virus responsible for the COVID-19 pandemic. The emergence of the new SARS-CoV-2 has been attributed to the possibility of evolutionary dynamics in the furin cleavage site (FCS) region. This study aimed to analyze the sequence of the FCS region in the spike protein of SARS-CoV-2 isolates that circulated in the Special Region of Yogyakarta and Central Java provinces in Indonesia. The RNA solution extracted from nasopharyngeal swab samples of confirmed COVID-19 patients were used and subjected to cDNA synthesis, PCR amplification, sequencing, and analysis of the FCS region. The sequence data from GISAID were also retrieved for further genome analysis. This study included 52 FCS region sequences. Several mutations were identified in the FCS region, i.e., D614G, Q675H, Q677H, S680P, and silent mutation in 235.57 C > T. The most important mutation in the FCS region is D614G. This finding indicated the G614 variant was circulating from May 2020 in those two provinces. Eventually, the G614 variant totally replaced the D614 variant from September 2020. All Indonesian SARS-CoV-2 isolates during this study and those deposited in GISAID showed the formation of five clade clusters from the FCS region, in which the D614 variant is in one specific cluster, and the G614 variant is dispersed into four clusters. The data indicated there is evolutionary advantage of the D614G mutation in the FCS region of the spike protein of SARS-CoV-2 circulating in the Special Region of Yogyakarta and Central Java provinces in Indonesia.
Keywords: D614G mutation; Furin cleavage site; SARS-CoV-2; Spike protein.
© 2022. The Author(s), under exclusive licence to Springer Nature Switzerland AG.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Begum F, Banerjee AK, Ray U (2020) Mutation hot spots in spike protein of SARS-CoV-2 virus. Preprints Available from: 10.20944/preprints202004.0281.v2. Accessed 15 Sept 2020
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

