CRISPR-Cas9 Gene Therapy for Duchenne Muscular Dystrophy
- PMID: 35165856
- PMCID: PMC9294086
- DOI: 10.1007/s13311-022-01197-9
CRISPR-Cas9 Gene Therapy for Duchenne Muscular Dystrophy
Abstract
Discovery of the CRISPR-Cas (clustered regularly interspaced short palindromic repeat, CRISPR-associated) system a decade ago has opened new possibilities in the field of precision medicine. CRISPR-Cas was initially identified in bacteria and archaea to play a protective role against foreign genetic elements during viral infections. The application of this technique for the correction of different mutations found in the Duchenne muscular dystrophy (DMD) gene led to the development of several potential therapeutic approaches for DMD patients. The mutations responsible for Duchenne muscular dystrophy mainly include exon deletions (70% of patients) and point mutations (about 30% of patients). The CRISPR-Cas 9 technology is becoming increasingly precise and is acquiring diverse functions through novel innovations such as base editing and prime editing. However, questions remain about its translation to the clinic. Current research addressing off-target editing, efficient muscle-specific delivery, immune response to nucleases, and vector challenges may eventually lead to the clinical use of the CRISPR-Cas9 technology. In this review, we present recent CRISPR-Cas9 strategies to restore dystrophin expression in vitro and in animal models of DMD.
Keywords: CRISPR-Cas; DMD gene; Duchenne muscular dystrophy; Dystrophin; Gene therapy.
© 2022. The American Society for Experimental NeuroTherapeutics, Inc.
Figures

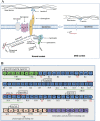

References
-
- Jansen R, van Embden JDA, Gaastra W, et al. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–1575. - PubMed
-
- Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. - PubMed
-
- Bolotin A, Quinquis B, Sorokin A, et al. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. - PubMed
-
- Pourcel C, Salvignol G, Vergnaud GY. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

