Neutralizing antibody responses elicited by SARS-CoV-2 mRNA vaccination wane over time and are boosted by breakthrough infection
- PMID: 35166573
- PMCID: PMC8939766
- DOI: 10.1126/scitranslmed.abn8057
Neutralizing antibody responses elicited by SARS-CoV-2 mRNA vaccination wane over time and are boosted by breakthrough infection
Abstract
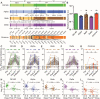
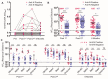
The waning efficacy of SARS-CoV-2 vaccines, combined with the continued emergence of variants resistant to vaccine-induced immunity, has reignited debate over the need for booster vaccine doses. To address this, we examined the neutralizing antibody response against the spike protein of five major SARS-CoV-2 variants, D614G, Alpha (B.1.1.7), Beta (B.1.351), Delta (B.1.617.2), and Omicron (B.1.1.529), in health care workers (HCWs) vaccinated with SARS-CoV-2 mRNA vaccines. Serum samples were collected before vaccination, 3 weeks after first vaccination, 1 month after second vaccination, and 6 months after second vaccination. Minimal neutralizing antibody titers were detected against Omicron pseudovirus at all four time points, including for most patients who had SARS-CoV-2 breakthrough infections. Neutralizing antibody titers against all other variant spike protein-bearing pseudoviruses declined markedly from 1 to 6 months after the second mRNA vaccine dose, although SARS-CoV-2 infection boosted vaccine responses. In addition, mRNA-1273-vaccinated HCWs exhibited about twofold higher neutralizing antibody titers than BNT162b2-vaccinated HCWs. Together, these results demonstrate possible waning of antibody-mediated protection against SARS-CoV-2 variants that is dependent on prior infection status and the mRNA vaccine received. They also show that the Omicron variant spike protein can almost completely escape from neutralizing antibodies elicited in recipients of only two mRNA vaccine doses.
Figures

References
-
- World Health Organization. COVID-19 weekly epidemiological update, 18 January 2022. (2021).
-
- Plante J. A., Liu Y., Liu J., Xia H., Johnson B. A., Lokugamage K. G., Zhang X., Muruato A. E., Zou J., Fontes-Garfias C. R., Mirchandani D., Scharton D., Bilello J. P., Ku Z., An Z., Kalveram B., Freiberg A. N., Menachery V. D., Xie X., Plante K. S., Weaver S. C., Shi P.-Y., Spike mutation D614G alters SARS-CoV-2 fitness. Nature 592, 116–121 (2021). 10.1038/s41586-020-2895-3 - DOI - PMC - PubMed
-
- Korber B., Fischer W. M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E. E., Bhattacharya T., Foley B., Hastie K. M., Parker M. D., Partridge D. G., Evans C. M., Freeman T. M., de Silva T. I., McDanal C., Perez L. G., Tang H., Moon-Walker A., Whelan S. P., LaBranche C. C., Saphire E. O., Montefiori D. C.; Sheffield COVID-19 Genomics Group , Tracking changes in SARS-CoV-2 Spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell 182, 812–827.e19 (2020). 10.1016/j.cell.2020.06.043 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

