Transcriptional analysis of lung fibroblasts identifies PIM1 signaling as a driver of aging-associated persistent fibrosis
- PMID: 35167499
- PMCID: PMC8986080
- DOI: 10.1172/jci.insight.153672
Transcriptional analysis of lung fibroblasts identifies PIM1 signaling as a driver of aging-associated persistent fibrosis
Abstract
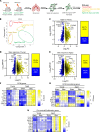
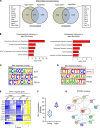
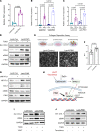
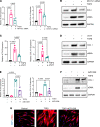
Idiopathic pulmonary fibrosis (IPF) is an aging-associated disease characterized by myofibroblast accumulation and progressive lung scarring. To identify transcriptional gene programs driving persistent lung fibrosis in aging, we performed RNA-Seq on lung fibroblasts isolated from young and aged mice during the early resolution phase after bleomycin injury. We discovered that, relative to injured young fibroblasts, injured aged fibroblasts exhibited a profibrotic state characterized by elevated expression of genes implicated in inflammation, matrix remodeling, and cell survival. We identified the proviral integration site for Moloney murine leukemia virus 1 (PIM1) and its target nuclear factor of activated T cells-1 (NFATc1) as putative drivers of the sustained profibrotic gene signatures in injured aged fibroblasts. PIM1 and NFATc1 transcripts were enriched in a pathogenic fibroblast population recently discovered in IPF lungs, and their protein expression was abundant in fibroblastic foci. Overexpression of PIM1 in normal human lung fibroblasts potentiated their fibrogenic activation, and this effect was attenuated by NFATc1 inhibition. Pharmacological inhibition of PIM1 attenuated IPF fibroblast activation and sensitized them to apoptotic stimuli. Interruption of PIM1 signaling in IPF lung explants ex vivo inhibited prosurvival gene expression and collagen secretion, suggesting that targeting this pathway may represent a therapeutic strategy to block IPF progression.
Keywords: Aging; Fibrosis; Pulmonology.
Conflict of interest statement
Figures

References
-
- Selman M, et al. Aging and pulmonary fibrosis. Rev Invest Clin. 2016;68(2):75–83. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

