Public human microbiome data are dominated by highly developed countries
- PMID: 35167588
- PMCID: PMC8846514
- DOI: 10.1371/journal.pbio.3001536
Public human microbiome data are dominated by highly developed countries
Abstract
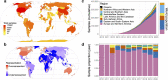
The importance of sampling from globally representative populations has been well established in human genomics. In human microbiome research, however, we lack a full understanding of the global distribution of sampling in research studies. This information is crucial to better understand global patterns of microbiome-associated diseases and to extend the health benefits of this research to all populations. Here, we analyze the country of origin of all 444,829 human microbiome samples that are available from the world's 3 largest genomic data repositories, including the Sequence Read Archive (SRA). The samples are from 2,592 studies of 19 body sites, including 220,017 samples of the gut microbiome. We show that more than 71% of samples with a known origin come from Europe, the United States, and Canada, including 46.8% from the US alone, despite the country representing only 4.3% of the global population. We also find that central and southern Asia is the most underrepresented region: Countries such as India, Pakistan, and Bangladesh account for more than a quarter of the world population but make up only 1.8% of human microbiome samples. These results demonstrate a critical need to ensure more global representation of participants in microbiome studies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Kaplan RC, Wang Z, Usyk M, Sotres-Alvarez D, Daviglus ML, Schneiderman N, et al. Gut microbiome composition in the Hispanic Community Health Study/Study of Latinos is shaped by geographic relocation, environmental factors, and obesity. Genome Biol. 2019;20:219. doi: 10.1186/s13059-019-1831-z - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

