Thirty years of resistance: Zig-zag through the plant immune system
- PMID: 35167697
- PMCID: PMC9048904
- DOI: 10.1093/plcell/koac041
Thirty years of resistance: Zig-zag through the plant immune system
Abstract
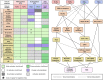
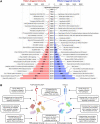
Understanding the plant immune system is crucial for using genetics to protect crops from diseases. Plants resist pathogens via a two-tiered innate immune detection-and-response system. The first plant Resistance (R) gene was cloned in 1992 . Since then, many cell-surface pattern recognition receptors (PRRs) have been identified, and R genes that encode intracellular nucleotide-binding leucine-rich repeat receptors (NLRs) have been cloned. Here, we provide a list of characterized PRRs and NLRs. In addition to immune receptors, many components of immune signaling networks were discovered over the last 30 years. We review the signaling pathways, physiological responses, and molecular regulation of both PRR- and NLR-mediated immunity. Recent studies have reinforced the importance of interactions between the two immune systems. We provide an overview of interactions between PRR- and NLR-mediated immunity, highlighting challenges and perspectives for future research.
© The Author(s) 2022. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Figures






References
-
- Adachi H, Derevnina L, Kamoun S (2019b) NLR singletons, pairs, and networks: evolution, assembly, and regulation of the intracellular immunoreceptor circuitry of plants. Curr Opin Plant Biol 50: 121–131 - PubMed
-
- Albert I, Albert M, Feiler C, Imkampe J, Brancato C, Raaymakers TM, Oome S, Wallmeroth N, Zhang H, Hedrich R, et al. (2015) An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat Plants 1: 15140. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

