Granger Causality Analysis of Chignolin Folding
- PMID: 35167755
- PMCID: PMC8908741
- DOI: 10.1021/acs.jctc.1c00945
Granger Causality Analysis of Chignolin Folding
Abstract
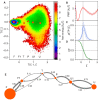
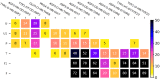
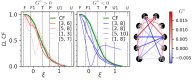
Constantly advancing computer simulations of biomolecules provide huge amounts of data that are difficult to interpret. In particular, obtaining insights into functional aspects of macromolecular dynamics, often related to cascades of transient events, calls for methodologies that depart from the well-grounded framework of equilibrium statistical physics. One of the approaches toward the analysis of complex temporal data which has found applications in the fields of neuroscience and econometrics is Granger causality analysis. It allows determining which components of multidimensional time series are most influential for the evolution of the entire system, thus providing insights into causal relations within the dynamic structure of interest. In this work, we apply Granger analysis to a long molecular dynamics trajectory depicting repetitive folding and unfolding of a mini β-hairpin protein, CLN025. We find objective, quantitative evidence indicating that rearrangements within the hairpin turn region are determinant for protein folding and unfolding. On the contrary, interactions between hairpin arms score low on the causality scale. Taken together, these findings clearly favor the concept of zipperlike folding, which is one of two postulated β-hairpin folding mechanisms. More importantly, the results demonstrate the possibility of a conclusive application of Granger causality analysis to a biomolecular system.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

