Structural and functional insights into the inhibition of human voltage-gated sodium channels by μ-conotoxin KIIIA disulfide isomers
- PMID: 35167877
- PMCID: PMC8927997
- DOI: 10.1016/j.jbc.2022.101728
Structural and functional insights into the inhibition of human voltage-gated sodium channels by μ-conotoxin KIIIA disulfide isomers
Abstract
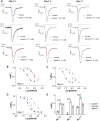
μ-Conotoxins are components of cone snail venom, well-known for their analgesic activity through potent inhibition of voltage-gated sodium channel (NaV) subtypes, including NaV1.7. These small, disulfide-rich peptides are typically stabilized by three disulfide bonds arranged in a 'native' CysI-CysIV, CysII-CysV, CysIII-CysVI pattern of disulfide connectivity. However, μ-conotoxin KIIIA, the smallest and most studied μ-conotoxin with inhibitory activity at NaV1.7, forms two distinct disulfide bond isomers during thermodynamic oxidative folding, including Isomer 1 (CysI-CysV, CysII-CysIV, CysIII-CysVI) and Isomer 2 (CysI-CysVI, CysII-CysIV, CysIII-CysV), but not the native μ-conotoxin arrangement. To date, there has been no study on the structure and activity of KIIIA comprising the native μ-conotoxin disulfide bond arrangement. Here, we evaluated the synthesis, potency, sodium channel subtype selectivity, and 3D structure of the three isomers of KIIIA. Using a regioselective disulfide bond-forming strategy, we synthetically produced the three μ-conotoxin KIIIA isomers displaying distinct bioactivity and NaV subtype selectivity across human NaV channel subtypes 1.2, 1.4, and 1.7. We show that Isomer 1 inhibits NaV subtypes with a rank order of potency of NaV1.4 > 1.2 > 1.7 and Isomer 2 in the order of NaV1.4≈1.2 > 1.7, while the native isomer inhibited NaV1.4 > 1.7≈1.2. The three KIIIA isomers were further evaluated by NMR solution structure analysis and molecular docking with hNaV1.2. Our study highlights the importance of investigating alternate disulfide isomers, as disulfide connectivity affects not only the overall structure of the peptides but also the potency and subtype selectivity of μ-conotoxins targeting therapeutically relevant NaV subtypes.
Keywords: KIIIA; Na(V)1.7; disulfide bond isomers; voltage-gated sodium channels; μ-conotoxin.
Copyright © 2022. Published by Elsevier Inc.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures



References
-
- Dib-Hajj S.D., Black J.A., Waxman S.G. Voltage-gated sodium channels: Therapeutic targets for pain. Pain Med. 2009;10:1260–1269. - PubMed
-
- Minett M.S., Pereira V., Sikandar S., Matsuyama A., Lolignier S., Kanellopoulos A.H., Mancini F., Iannetti G.D., Bogdanov Y.D., Santana-Varela S., Millet Q., Baskozos G., MacAllister R., Cox J.J., Zhao J., et al. Endogenous opioids contribute to insensitivity to pain in humans and mice lacking sodium channel NaV1.7. Nat. Commun. 2015;6:1–16. - PMC - PubMed
-
- Dib-Hajj S.D., Black J.A., Waxman S.G. NaV1.9: A sodium channel linked to human pain. Nat. Rev. Neurosci. 2015;16:511–519. - PubMed
-
- Osteen J.D., Herzig V., Gilchrist J., Emrick J.J., Zhang C., Wang X., Castro J., Garcia-Caraballo S., Grundy L., Rychkov G.Y., Weyer A.D., Dekan Z., Undheim E.A., Alewood P., Stucky C.L., et al. Selective spider toxins reveal a role for the NaV1.1 channel in mechanical pain. Nature. 2016;534:494–499. - PMC - PubMed
-
- Vetter I., Deuis J.R., Mueller A., Israel M.R., Starobova H., Zhang A., Rash L.D., Mobli M. NaV1.7 as a pain target - from gene to pharmacology. Pharmacol. Ther. 2017;172:73–100. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

