NEDD4 degrades TUSC2 to promote glioblastoma progression
- PMID: 35167936
- PMCID: PMC8920049
- DOI: 10.1016/j.canlet.2022.01.029
NEDD4 degrades TUSC2 to promote glioblastoma progression
Abstract
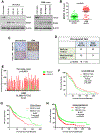
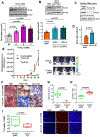
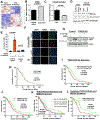
Whether tumor suppressor candidate 2 (TUSC2) plays an important role in glioblastoma (GBM) progression is largely unknown. Whether TUSC2 undergoes polyubiquitination is unknown. Herein, we report that TUSC2 protein expression is reduced/lost in GBM compared to normal brain due to protein destabilization; TUSC2 mRNA is equally expressed in both tissues. NEDD4 E3 ubiquitin ligase polyubiquitinates TUSC2 at residue K71, and the TUSC2-K71R mutant is resistant to NEDD4-mediated proteasomal degradation. Analysis of GBM specimens showed NEDD4 protein is highly expressed in GBM and the level is inversely correlated with TUSC2 protein levels. Furthermore, TUSC2 restoration induces apoptosis and inhibits patient-derived glioma stem cells (PD-GSCs) in vitro and in vivo. Conversely, TUSC2-knockout promotes PD-GSCs in vitro and in vivo. RNA-Seq analysis and subsequent validations showed GBM cells with TUSC2-knockout expressed increased Bcl-xL and were more resistant to apoptosis induced by a Bcl-xL-specific BH3 mimetic. A TUSC2-knockout gene signature created from the RNA-seq data predicts poor patient survival. Together, these findings establish that NEDD4-mediated polyubiquitination is a novel mechanism for TUSC2 degradation in GBM and that TUSC2 loss promotes GBM progression in part through Bcl-xL upregulation.
Keywords: Glioblastoma; Glioma stem cells; NEDD4; TUSC2; Tumor suppressor.
Copyright © 2022 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Interest Statement
Authors declare no conflict of interests.
Figures




References
-
- Omuro A, DeAngelis LM, Glioblastoma and other malignant gliomas: A clinical review, JAMA, 310 (2013) 1842–1850. - PubMed
-
- Anjum K, Shagufta BI, Abbas SO, Patel S, Khan I, Shah SAA, et al. , Current status and future therapeutic perspectives of glioblastoma multiforme (GBM) therapy: A review, Biomedicine and Parmacotherapy, 92 (2017) 10. - PubMed
-
- Brennan CW, Verhaak RGW, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, Beroukhim R, Bernard B, Wu C-J, Genovese G, Shmulevich I, Barnholtz-Sloan J, Zou L, Vegesna R, Shukla SA, Ciriello G, Yung WK, Zhang W, Sougnez C, Mikkelsen T, Aldape K, Bigner DD, Van Meir EG, Prados M, Sloan A, Black KL, Eschbacher J, Finocchiaro G, Friedman W, Andrews DW, Guha A, Iacocca M, O’Neill BP, Foltz G, Myers J, Weisenberger DJ, Penny R, Kucherlapati R, Perou CM, Hayes DN, Gibbs R, Marra M, Mills GB, Lander E, Spellman P, Wilson R, Sander C, Weinstein J, Meyerson M, Gabriel S, Laird PW, Haussler D, Getz G, Chin L, The Somatic Genomic Landscape of Glioblastoma, Cell, 155 (2013) 462–477. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

