Diffusion time dependence, power-law scaling, and exchange in gray matter
- PMID: 35168088
- PMCID: PMC8961002
- DOI: 10.1016/j.neuroimage.2022.118976
Diffusion time dependence, power-law scaling, and exchange in gray matter
Abstract
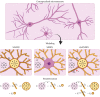
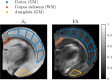
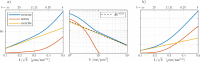
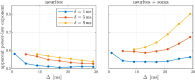
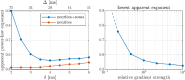
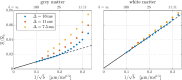
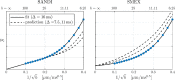
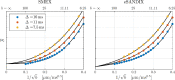
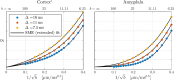
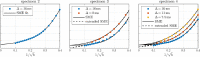
Characterizing neural tissue microstructure is a critical goal for future neuroimaging. Diffusion MRI (dMRI) provides contrasts that reflect diffusing spins' interactions with myriad microstructural features of biological systems. However, the specificity of dMRI remains limited due to the ambiguity of its signals vis-à-vis the underlying microstructure. To improve specificity, biophysical models of white matter (WM) typically express dMRI signals according to the Standard Model (SM) and have more recently in gray matter (GM) taken spherical compartments into account (the SANDI model) in attempts to represent cell soma. The validity of the assumptions underlying these models, however, remains largely undetermined, especially in GM. To validate these assumptions experimentally, observing their unique, functional properties, such as the b-1/2 power-law associated with one-dimensional diffusion, has emerged as a fruitful strategy. The absence of this signature in GM, in turn, has been explained by neurite water exchange, non-linear morphology, and/or by obscuring soma signal contributions. Here, we present diffusion simulations in realistic neurons demonstrating that curvature and branching does not destroy the stick power-law behavior in impermeable neurites, but also that their signal is drowned by the soma signal under typical experimental conditions. Nevertheless, by studying the GM dMRI signal's behavior as a function of diffusion weighting as well as time, we identify an attainable experimental regime in which the neurite signal dominates. Furthermore, we find that exchange-driven time dependence produces a signal behavior opposite to that which would be expected from restricted diffusion, thereby providing a functional signature that disambiguates the two effects. We present data from dMRI experiments in ex vivo rat brain at ultrahigh field of 16.4T and observe a time dependence that is consistent with substantial exchange but also with a GM stick power-law. The first finding suggests significant water exchange between neurites and the extracellular space while the second suggests a small sub-population of impermeable neurites. To quantify these observations, we harness the Kärger exchange model and incorporate the corresponding signal time dependence in the SM and SANDI models.
Keywords: Diffusion MRI; Exchange; Gray matter; Microstructure; Soma and neurite density imaging; Standard model.
Copyright © 2022. Published by Elsevier Inc.
Figures


Similar articles
-
Neurite Exchange Imaging (NEXI): A minimal model of diffusion in gray matter with inter-compartment water exchange.Neuroimage. 2022 Aug 1;256:119277. doi: 10.1016/j.neuroimage.2022.119277. Epub 2022 May 3. Neuroimage. 2022. PMID: 35523369 Free PMC article.
-
Soma and Neurite Density MRI (SANDI) of the in-vivo mouse brain and comparison with the Allen Brain Atlas.Neuroimage. 2022 Jul 1;254:119135. doi: 10.1016/j.neuroimage.2022.119135. Epub 2022 Mar 23. Neuroimage. 2022. PMID: 35339686
-
Mapping tissue microstructure across the human brain on a clinical scanner with soma and neurite density image metrics.Hum Brain Mapp. 2023 Sep;44(13):4792-4811. doi: 10.1002/hbm.26416. Epub 2023 Jul 17. Hum Brain Mapp. 2023. PMID: 37461286 Free PMC article.
-
Probing brain tissue microstructure with MRI: principles, challenges, and the role of multidimensional diffusion-relaxation encoding.Neuroimage. 2023 Nov 15;282:120338. doi: 10.1016/j.neuroimage.2023.120338. Epub 2023 Aug 19. Neuroimage. 2023. PMID: 37598814 Review.
-
In Vivo Imaging of Gray Matter Microstructure in Major Psychiatric Disorders: Opportunities for Clinical Translation.Biol Psychiatry Cogn Neurosci Neuroimaging. 2020 Sep;5(9):855-864. doi: 10.1016/j.bpsc.2020.03.003. Epub 2020 Mar 25. Biol Psychiatry Cogn Neurosci Neuroimaging. 2020. PMID: 32381477 Review.
Cited by
-
Surface-based Analyses of Diffusional Kurtosis Imaging in Amyotrophic Lateral Sclerosis: Relationship with Onset Subtypes.Magn Reson Med Sci. 2025 Jan 1;24(1):122-132. doi: 10.2463/mrms.mp.2023-0138. Epub 2024 May 29. Magn Reson Med Sci. 2025. PMID: 38296522 Free PMC article.
-
Temporal Diffusion Ratio (TDR) for imaging restricted diffusion: Optimisation and pre-clinical demonstration.Neuroimage. 2023 Apr 1;269:119930. doi: 10.1016/j.neuroimage.2023.119930. Epub 2023 Feb 5. Neuroimage. 2023. PMID: 36750150 Free PMC article.
-
Detection of grey matter microstructural substrates of neurodegeneration in multiple sclerosis.Brain Commun. 2023 May 24;5(3):fcad153. doi: 10.1093/braincomms/fcad153. eCollection 2023. Brain Commun. 2023. PMID: 37274832 Free PMC article.
-
Mean Kärger Model Water Exchange Rate in Brain.Imaging Neurosci (Camb). 2024;2:imag-2-00335. doi: 10.1162/imag_a_00335. Epub 2024 Oct 25. Imaging Neurosci (Camb). 2024. PMID: 40534623 Free PMC article.
-
Human gray matter microstructure mapped using neurite exchange imaging (NEXI) on a clinical scanner.Imaging Neurosci (Camb). 2025 Jun 12;3:IMAG.a.32. doi: 10.1162/IMAG.a.32. eCollection 2025. Imaging Neurosci (Camb). 2025. PMID: 40800941 Free PMC article.
References
-
- Ascoli G.A., Donohue D.E., Halavi M. NeuroMorpho.Org: a central resource for neuronal morphologies. J. Neurosci. 2007 doi: 10.1523/JNEUROSCI.2055-07.2007. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

