Efficiently sparse listing of classes of optimal cophylogeny reconciliations
- PMID: 35168648
- PMCID: PMC8845303
- DOI: 10.1186/s13015-022-00206-y
Efficiently sparse listing of classes of optimal cophylogeny reconciliations
Abstract
Background: Cophylogeny reconciliation is a powerful method for analyzing host-parasite (or host-symbiont) co-evolution. It models co-evolution as an optimization problem where the set of all optimal solutions may represent different biological scenarios which thus need to be analyzed separately. Despite the significant research done in the area, few approaches have addressed the problem of helping the biologist deal with the often huge space of optimal solutions.
Results: In this paper, we propose a new approach to tackle this problem. We introduce three different criteria under which two solutions may be considered biologically equivalent, and then we propose polynomial-delay algorithms that enumerate only one representative per equivalence class (without listing all the solutions).
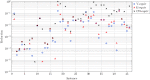
Conclusions: Our results are of both theoretical and practical importance. Indeed, as shown by the experiments, we are able to significantly reduce the space of optimal solutions while still maintaining important biological information about the whole space.
Keywords: Cophylogeny; Dynamic programming; Enumeration; Equivalence relation.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
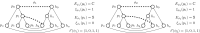

References
-
- Pennington PM, Messenger LA, Reina J, Juárez JG, Lawrence GG, Dotson EM, Llewellyn MS, Cordón-Rosales C. The chagas disease domestic transmission cycle in guatemala: parasite-vector switches and lack of mitochondrial co-diversification between triatoma dimidiata and trypanosoma cruzi subpopulations suggest non-vectorial parasite dispersal across the motagua valley. Acta Tropica. 2015;151:80–7. doi: 10.1016/j.actatropica.2015.07.014. - DOI - PubMed
LinkOut - more resources
Full Text Sources

