A population study of clinically actionable genetic variation affecting drug response from the Middle East
- PMID: 35169154
- PMCID: PMC8847489
- DOI: 10.1038/s41525-022-00281-5
A population study of clinically actionable genetic variation affecting drug response from the Middle East
Abstract
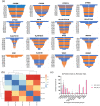
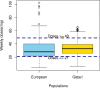
Clinical implementation of pharmacogenomics will help in personalizing drug prescriptions and alleviate the personal and financial burden due to inefficacy and adverse reactions to drugs. However, such implementation is lagging in many parts of the world, including the Middle East, mainly due to the lack of data on the distribution of actionable pharmacogenomic variation in these ethnicities. We analyzed 6,045 whole genomes from the Qatari population for the distribution of allele frequencies of 2,629 variants in 1,026 genes known to affect 559 drugs or classes of drugs. We also performed a focused analysis of genotypes or diplotypes of 15 genes affecting 46 drugs, which have guidelines for clinical implementation and predicted their phenotypic impact. The allele frequencies of 1,320 variants in 703 genes affecting 299 drugs or class of drugs were significantly different between the Qatari population and other world populations. On average, Qataris carry 3.6 actionable genotypes/diplotypes, affecting 13 drugs with guidelines for clinical implementation, and 99.5% of the individuals had at least one clinically actionable genotype/diplotype. Increased risk of simvastatin-induced myopathy could be predicted in ~32% of Qataris from the diplotypes of SLCO1B1, which is higher compared to many other populations, while fewer Qataris may need tacrolimus dosage adjustments for achieving immunosuppression based on the CYP3A5 diplotypes compared to other world populations. Distinct distribution of actionable pharmacogenomic variation was also observed among the Qatari subpopulations. Our comprehensive study of the distribution of actionable genetic variation affecting drugs in a Middle Eastern population has potential implications for preemptive pharmacogenomic implementation in the region and beyond.
© 2022. The Author(s).
Conflict of interest statement
MP receives research funding from various organisations including the UK MRC and NIHR. He has also received partnership funding for the following: MRC Clinical Pharmacology Training Scheme (co-funded by MRC and Roche, UCB, Eli Lilly and Novartis); a PhD studentship jointly funded by EPSRC and Astra Zeneca; and grant funding from Vistagen Therapeutics. He has also unrestricted educational grant support for the UK Pharmacogenetics and Stratified Medicine Network from Bristol-Myers Squibb and UCB. He has developed an HLA genotyping panel with MC Diagnostics, but does not benefit financially from this. He is part of the IMI Consortium ARDAT (
Figures
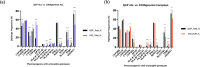
References
-
- Pirmohamed M. Personalized pharmacogenomics: predicting efficacy and adverse drug reactions. Annu. Rev. Genomics Hum. Genet. 2014;15:349–370. - PubMed
-
- Caspar SM, Schneider T, Stoll P, Meienberg J, Matyas G. Potential of whole-genome sequencing-based pharmacogenetic profiling. Pharmacogenomics. 2021;22:177–190. - PubMed
LinkOut - more resources
Full Text Sources

