Non-covalent SARS-CoV-2 Mpro inhibitors developed from in silico screen hits
- PMID: 35169179
- PMCID: PMC8847420
- DOI: 10.1038/s41598-022-06306-4
Non-covalent SARS-CoV-2 Mpro inhibitors developed from in silico screen hits
Abstract
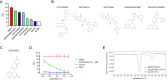
Mpro, the main protease of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is essential for the viral life cycle. Accordingly, several groups have performed in silico screens to identify Mpro inhibitors that might be used to treat SARS-CoV-2 infections. We selected more than five hundred compounds from the top-ranking hits of two very large in silico screens for on-demand synthesis. We then examined whether these compounds could bind to Mpro and inhibit its protease activity. Two interesting chemotypes were identified, which were further evaluated by characterizing an additional five hundred synthesis on-demand analogues. The compounds of the first chemotype denatured Mpro and were considered not useful for further development. The compounds of the second chemotype bound to and enhanced the melting temperature of Mpro. The most active compound from this chemotype inhibited Mpro in vitro with an IC50 value of 1 μM and suppressed replication of the SARS-CoV-2 virus in tissue culture cells. Its mode of binding to Mpro was determined by X-ray crystallography, revealing that it is a non-covalent inhibitor. We propose that the inhibitors described here could form the basis for medicinal chemistry efforts that could lead to the development of clinically relevant inhibitors.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

