Niche partitioning of the ubiquitous and ecologically relevant NS5 marine group
- PMID: 35169264
- PMCID: PMC9122927
- DOI: 10.1038/s41396-022-01209-8
Niche partitioning of the ubiquitous and ecologically relevant NS5 marine group
Abstract
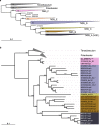
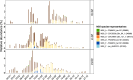
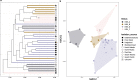
Niche concept is a core tenet of ecology that has recently been applied in marine microbial research to describe the partitioning of taxa based either on adaptations to specific conditions across environments or on adaptations to specialised substrates. In this study, we combine spatiotemporal dynamics and predicted substrate utilisation to describe species-level niche partitioning within the NS5 Marine Group. Despite NS5 representing one of the most abundant marine flavobacterial clades from across the world's oceans, our knowledge on their phylogenetic diversity and ecological functions is limited. Using novel and database-derived 16S rRNA gene and ribosomal protein sequences, we delineate the NS5 into 35 distinct species-level clusters, contained within four novel candidate genera. One candidate species, "Arcticimaribacter forsetii AHE01FL", includes a novel cultured isolate, for which we provide a complete genome sequence-the first of an NS5-along with morphological insights using transmission electron microscopy. Assessing species' spatial distribution dynamics across the Tara Oceans dataset, we identify depth as a key influencing factor, with 32 species preferring surface waters, as well as distinct patterns in relation to temperature, oxygen and salinity. Each species harbours a unique substrate-degradation potential along with predicted substrates conserved at the genus-level, e.g. alginate in NS5_F. Successional dynamics were observed for three species in a time-series dataset, likely driven by specialised substrate adaptations. We propose that the ecological niche partitioning of NS5 species is mainly based on specific abiotic factors, which define the niche space, and substrate availability that drive the species-specific temporal dynamics.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Hutchinson GE. Concluding remarks. Cold Spring Harb Symp Quant Biol. 1957;22:415–27.
-
- Hutchinson GE. An introduction to population biology. New Haven, CT: Yale University Press; 1978.
-
- Larkin AA, Martiny AC. Microdiversity shapes the traits, niche space, and biogeography of microbial taxa. Environ Microbiol Rep. 2017;9:55–70. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

