Resolving taxonomic confusion: establishing the genus Phytobacter on the list of clinically relevant Enterobacteriaceae
- PMID: 35169969
- PMCID: PMC8934334
- DOI: 10.1007/s10096-022-04413-8
Resolving taxonomic confusion: establishing the genus Phytobacter on the list of clinically relevant Enterobacteriaceae
Abstract
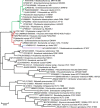
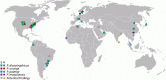
Although many clinically significant strains belonging to the family Enterobacteriaceae fall into a restricted number of genera and species, there is still a substantial number of isolates that elude this classification and for which proper identification remains challenging. With the current improvements in the field of genomics, it is not only possible to generate high-quality data to accurately identify individual nosocomial isolates at the species level and understand their pathogenic potential but also to analyse retrospectively the genome sequence databases to identify past recurrences of a specific organism, particularly those originally published under an incorrect or outdated taxonomy. We propose a general use of this approach to classify further clinically relevant taxa, i.e., Phytobacter spp., that have so far gone unrecognised due to unsatisfactory identification procedures in clinical diagnostics. Here, we present a genomics and literature-based approach to establish the importance of the genus Phytobacter as a clinically relevant member of the Enterobacteriaceae family.
Keywords: Genomics; Identification; Phytobacter diazotrophicus; Phytobacter palmae; Phytobacter ursingii; Taxonomy.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Skerman VBD, McGowan V, Sneath PHA. Approved lists of bacterial names. Int J Syst Bacteriol. 1980;30(1):225–420. - PubMed
-
- Adeolu M, Alnajar S, Naushad S, Gupta RS. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int J Syst Evol Microbiol. 2016;66:5575–5599. - PubMed
-
- Alnajar S, Gupta RS. Phylogenomics and comparative genomic studies delineate six main clades within the family Enterobacteriaceae and support the reclassification of several polyphyletic members of the family. Infect Genet Evol. 2017;54:108–127. - PubMed
-
- Rosselló-Móra R, Amann R. Past and future species definitions for Bacteria and Archaea. Syst Appl Microbiol. 2015;38:209–216. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

