Tomato CRABS CLAW paralogues interact with chromatin remodelling factors to mediate carpel development and floral determinacy
- PMID: 35170044
- PMCID: PMC9314824
- DOI: 10.1111/nph.18034
Tomato CRABS CLAW paralogues interact with chromatin remodelling factors to mediate carpel development and floral determinacy
Abstract
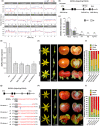
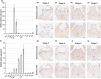
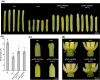
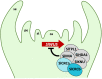
CRABS CLAW (CRC) orthologues play a crucial role in floral meristem (FM) determinacy and gynoecium formation across angiosperms, the key developmental processes for ensuring successful plant reproduction and crop production. However, the mechanisms behind CRC mediated FM termination are far from fully understood. Here, we addressed the functional characterization of tomato (Solanum lycopersicum) paralogous CRC genes. Using mapping-by-sequencing, RNA interference and CRISPR/Cas9 techniques, expression analyses, protein-protein interaction assays and Arabidopsis complementation experiments, we examined their potential roles in FM determinacy and carpel formation. We revealed that the incomplete penetrance and variable expressivity of the indeterminate carpel-inside-carpel phenotype observed in fruit iterative growth (fig) mutant plants are due to the lack of function of the S. lycopersicum CRC homologue SlCRCa. Furthermore, a detailed functional analysis of tomato CRC paralogues, SlCRCa and SlCRCb, allowed us to propose that they operate as positive regulators of FM determinacy by acting in a compensatory and partially redundant manner to safeguard the proper formation of flowers and fruits. Our results uncover for the first time the physical interaction of putative CRC orthologues with members of the chromatin remodelling complex that epigenetically represses WUSCHEL expression through histone deacetylation to ensure the proper termination of floral stem cell activity.
Keywords: CRABS CLAW (CRC); WUSCHEL (WUS); carpel development; floral meristem determinacy; fruit formation; incomplete penetrance; tomato (Solanum lycopersicum); variable expressivity.
© 2022 The Authors. New Phytologist © 2022 New Phytologist Foundation.
Figures



References
-
- Alvarez J, Smyth DR. 1999. CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS . Development 126: 2377–2386. - PubMed
-
- Bartholmes C, Hidalgo O, Gleissberg S. 2012. Evolution of the YABBY gene family with emphasis on the basal eudicot Eschscholzia californica (Papaveraceae). Plant Biology 14: 11–23. - PubMed
-
- Birchler JA, Johnson AF, Veitia RA. 2016. Kinetics genetics: incorporating the concept of genomic balance into an understanding of quantitative traits. Plant Science 245: 128–134. - PubMed
-
- Bollier N, Sicard A, Leblond J, Latrasse D, Gonzalez N, Gévaudant F, Benhamed M, Raynaud C, Lenhard M, Chevalier C et al. 2018. At‐MINI ZINC FINGER2 and Sl‐INHIBITOR OF MERISTEM ACTIVITY, a conserved missing link in the regulation of floral meristem termination in Arabidopsis and tomato. Plant cell 30: 83–100. - PMC - PubMed
-
- Bowman JL, Smyth DR. 1999. CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix‐loop‐helix domains. Development 126: 2387–2396. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

