Diagnostic yield of exome sequencing for prenatal diagnosis of fetal structural anomalies: A systematic review and meta-analysis
- PMID: 35170059
- PMCID: PMC9325531
- DOI: 10.1002/pd.6115
Diagnostic yield of exome sequencing for prenatal diagnosis of fetal structural anomalies: A systematic review and meta-analysis
Abstract
Objectives: We conducted a systematic review and meta-analysis to determine the diagnostic yield of exome sequencing (ES) for prenatal diagnosis of fetal structural anomalies, where karyotype/chromosomal microarray (CMA) is normal.
Methods: Following electronic searches of four databases, we included studies with ≥10 structurally abnormal fetuses undergoing ES or whole genome sequencing. The incremental diagnostic yield of ES over CMA/karyotype was calculated and pooled in a meta-analysis. Sub-group analyses investigated effects of case selection and fetal phenotype on diagnostic yield.
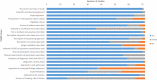
Results: We identified 72 reports from 66 studies, representing 4350 fetuses. The pooled incremental yield of ES was 31% (95% confidence interval (CI) 26%-36%, p < 0.0001). Diagnostic yield was significantly higher for cases pre-selected for likelihood of monogenic aetiology compared to unselected cases (42% vs. 15%, p < 0.0001). Diagnostic yield differed significantly between phenotypic sub-groups, ranging from 53% (95% CI 42%-63%, p < 0.0001) for isolated skeletal abnormalities, to 2% (95% CI 0%-5%, p = 0.04) for isolated increased nuchal translucency.
Conclusion: Prenatal ES provides a diagnosis in an additional 31% of structurally abnormal fetuses when CMA/karyotype is non-diagnostic. The expected diagnostic yield depends on the body system(s) affected and can be optimised by pre-selection of cases following multi-disciplinary review to determine that a monogenic cause is likely.
© 2022 The Authors. Prenatal Diagnosis published by John Wiley & Sons Ltd.
Conflict of interest statement
None of the authors have any conflict of interests to declare.
Figures






References
-
- Syngelaki A, Hammami A, Bower S, Zidere V, Akolekar R, Nicolaides KH. Diagnosis of fetal non‐chromosomal abnormalities on routine ultrasound examination at 11–13 weeks’ gestation. Ultrasound Obstet Gynecol. 2019;54(4):468‐476. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

