Long-Term Hepatitis B Virus Infection Induces Cytopathic Effects in Primary Human Hepatocytes, and Can Be Partially Reversed by Antiviral Therapy
- PMID: 35171034
- PMCID: PMC8849052
- DOI: 10.1128/spectrum.01328-21
Long-Term Hepatitis B Virus Infection Induces Cytopathic Effects in Primary Human Hepatocytes, and Can Be Partially Reversed by Antiviral Therapy
Abstract
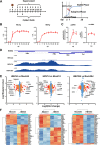
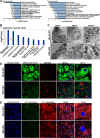
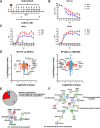
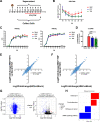
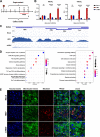
Chronic infection of hepatitis B virus (HBV) remains a major health burden worldwide. While the immune response has been recognized to play crucial roles in HBV pathogenesis, the direct cytopathic effects of HBV infection and replication on host hepatocytes and the HBV-host interactions are only partially defined due to limited culture systems. Here, based on our recently developed 5 chemical-cultured primary human hepatocytes (5C-PHHs) model that supports long-term HBV infection, we performed multiplexed quantitative analysis of temporal changes of host proteome and transcriptome on PHHs infected by HBV for up to 4 weeks. We showed that metabolic-, complement-, cytoskeleton-, mitochondrial-, and oxidation-related pathways were modulated at transcriptional or posttranscriptional levels during long-term HBV infection, which led to cytopathic effects and could be partially rescued by early, rather than late, nucleot(s)ide analog (NA) administration and could be significantly relieved by blocking viral antigens with RNA interference (RNAi). Overexpression screening of the dysregulated proteins identified a series of host factors that may contribute to pro- or anti-HBV responses of the infected hepatocytes. In conclusion, our results suggest that long-term HBV infection in primary human hepatocytes leads to cytopathic effects through remodeling the proteome and transcriptome and early antiviral treatment may reduce the extent of such effects, indicating a role of virological factors in HBV pathogenesis and a potential benefit of early administration of antiviral treatment. IMPORTANCE Global temporal quantitative proteomic and transcriptomic analysis using long-term hepatitis B virus (HBV)-infected primary human hepatocytes uncovered extensive remodeling of the host proteome and transcriptome and revealed cytopathic effects of long-term viral replication. Metabolic-, complement-, cytoskeleton-, mitochondrial-, and oxidation-related pathways were modulated at transcriptional or posttranscriptional levels, which could be partially rescued by early, rather than late, NA therapy and could be relieved by blocking viral antigens with RNAi. Overexpression screening identified a series of pro- or anti-HBV host factors. These data have deepened the understanding of the mechanisms of viral pathogenesis and HBV-host interactions in hepatocytes, with implications for therapeutic intervention.
Keywords: cytopathic effects; long-term HBV infection; nucleot(s)ide analogues; quantitative temporal proteomics; viral-host interaction.
Conflict of interest statement
The authors declare no conflict of interest.
All authors who have taken part in this study declare that we do not have anything to disclose regarding funding or conflict of interest with respect to the manuscript.
Figures


Similar articles
-
Analysis of host factor networks during hepatitis B virus infection in primary human hepatocytes.Virol J. 2024 Aug 1;21(1):170. doi: 10.1186/s12985-024-02446-3. Virol J. 2024. PMID: 39090742 Free PMC article.
-
Human stem cell-derived hepatocytes as a model for hepatitis B virus infection, spreading and virus-host interactions.J Hepatol. 2017 Mar;66(3):494-503. doi: 10.1016/j.jhep.2016.10.009. Epub 2016 Oct 14. J Hepatol. 2017. PMID: 27746336 Free PMC article.
-
HBV Bypasses the Innate Immune Response and Does Not Protect HCV From Antiviral Activity of Interferon.Gastroenterology. 2018 May;154(6):1791-1804.e22. doi: 10.1053/j.gastro.2018.01.044. Epub 2018 Feb 1. Gastroenterology. 2018. PMID: 29410097
-
Latest developments in the treatment of hepatitis B.Minerva Gastroenterol Dietol. 2016 Mar;62(1):88-102. Epub 2015 Oct 8. Minerva Gastroenterol Dietol. 2016. PMID: 26448309 Review.
-
HBV evolution and genetic variability: Impact on prevention, treatment and development of antivirals.Antiviral Res. 2021 Feb;186:104973. doi: 10.1016/j.antiviral.2020.104973. Epub 2020 Nov 6. Antiviral Res. 2021. PMID: 33166575 Review.
Cited by
-
Interactomics: Dozens of Viruses, Co-evolving With Humans, Including the Influenza A Virus, may Actively Distort Human Aging.Mol Biol Evol. 2023 Feb 3;40(2):msad012. doi: 10.1093/molbev/msad012. Mol Biol Evol. 2023. PMID: 36649176 Free PMC article.
-
Advances in multi-omics research on viral hepatitis.Front Microbiol. 2022 Sep 2;13:987324. doi: 10.3389/fmicb.2022.987324. eCollection 2022. Front Microbiol. 2022. PMID: 36118247 Free PMC article. Review.
-
A novel anti-HBV agent, E-CFCP, restores Hepatitis B virus (HBV)-induced senescence-associated cellular marker perturbation in human hepatocytes.Virus Res. 2023 May;329:199094. doi: 10.1016/j.virusres.2023.199094. Epub 2023 Mar 23. Virus Res. 2023. PMID: 36933835 Free PMC article.
-
Progress of mitochondrial and endoplasmic reticulum-associated signaling and its regulation of chronic liver disease by Chinese medicine.World J Hepatol. 2024 Apr 27;16(4):494-505. doi: 10.4254/wjh.v16.i4.494. World J Hepatol. 2024. PMID: 38689744 Free PMC article.
-
Host cytokine genetic polymorphisms in a selected population of persons living with hepatitis B virus infection in the central region of Ghana.BMC Gastroenterol. 2024 Oct 22;24(1):374. doi: 10.1186/s12876-024-03456-9. BMC Gastroenterol. 2024. PMID: 39434005 Free PMC article.
References
-
- Howell J, Pedrana A, Schroeder SE, Scott N, Aufegger L, Atun R, Baptista-Leite R, Hirnschall G, T Hoen E, Hutchinson SJ, Lazarus JV, Olufunmilayo L, Peck R, Sharma M, Sohn AH, Thompson A, Thursz M, Wilson D, Hellard M. 2021. A global investment framework for the elimination of hepatitis B. J Hepatol 74:535–549. doi:10.1016/j.jhep.2020.09.013. - DOI - PMC - PubMed
-
- Lau GKK, Piratvisuth T, Luo KX, Marcellin P, Thongsawat S, Cooksley G, Gane E, Fried MW, Chow WC, Paik SW, Chang WY, Berg T, Flisiak R, McCloud P, Pluck N, Peginterferon AAHC. 2005. Peginterferon alfa-2a, lamivudine, and the combination for HBeAg-positive chronic hepatitis B. N Engl J Med 352:2682–2695. doi:10.1056/NEJMoa043470. - DOI - PubMed
-
- Nakamoto Y, Guidotti LG, Pasquetto V, Schreiber RD, Chisari FV. 1997. Differential target cell sensitivity to CTL-activated death pathways in hepatitis B virus transgenic mice. J Immunol 158:5692–5697. - PubMed
-
- Maini MK, Boni C, Lee CK, Larrubia JR, Reignat S, Ogg GS, King AS, Herberg J, Gilson R, Alisa A, Williams R, Vergani D, Naoumov NV, Ferrari C, Bertoletti A. 2000. The role of virus-specific CD8(+) cells in liver damage and viral control during persistent hepatitis B virus infection. J Exp Med 191:1269–1280. doi:10.1084/jem.191.8.1269. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

