Monitoring of Dynamic Changes and Clonal Evolution in Circulating Tumor DNA From Patients With IDH-Mutated Cholangiocarcinoma Treated With Isocitrate Dehydrogenase Inhibitors
- PMID: 35171660
- PMCID: PMC8865526
- DOI: 10.1200/PO.21.00197
Monitoring of Dynamic Changes and Clonal Evolution in Circulating Tumor DNA From Patients With IDH-Mutated Cholangiocarcinoma Treated With Isocitrate Dehydrogenase Inhibitors
Abstract
Purpose: IDH mutations occur in about 30% of patients with cholangiocarcinoma. Analysis of mutations in circulating tumor DNA (ctDNA) can be performed by droplet digital polymerase chain reaction (ddPCR). The analysis of ctDNA is a feasible approach to detect IDH mutations.
Methods: We isolated ctDNA from the blood of patients with IDH-mutated advanced cholangiocarcinoma collected at baseline, on therapy, and at progression to isocitrate dehydrogenase (IDH) inhibitors.
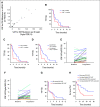
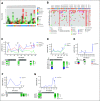
Results: Of 31 patients with IDH1R132 (n = 26) or IDH2R172 mutations (n = 5) in the tumor, IDH mutations were detected in 84% of ctDNA samples analyzed by ddPCR and in 83% of ctDNA samples analyzed by next-generation sequencing (NGS). Patients with a low variant allele frequency of ctDNA detected by NGS at baseline had a longer median time to treatment failure compared to patients with high variant allele frequency of ctDNA (3.6 v 1.5 months; P = .008). Patients with a decrease in IDH-mutated ctDNA on therapy by ddPCR compared with no change/increase had a trend to a longer median survival (P = .07). Most frequent emergent alterations in ctDNA by NGS at progression were ARID1A (n = 3) and TP53 mutations (n = 3).
Conclusion: Detection of IDH mutations in ctDNA in patients with advanced cholangiocarcinoma is feasible, and dynamic changes in ctDNA can correspond with the clinical course and clonal evolution.
Conflict of interest statement
Figures
References
-
- Fujii T, Khawaja MR, DiNardo CD, et al. Targeting isocitrate dehydrogenase (IDH) in cancer Discov Med 21373–3802016 - PubMed
-
- Javle M, Bekaii-Saab T, Jain A, et al. Biliary cancer: Utility of next-generation sequencing for clinical management Cancer 1223838–38472016 - PubMed
-
- DiNardo CD, Stein EM, de Botton S, et al. Durable remissions with ivosidenib in IDH1-mutated relapsed or refractory AML N Engl J Med 3782386–23982018 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

