A balanced Oct4 interactome is crucial for maintaining pluripotency
- PMID: 35171666
- PMCID: PMC8849292
- DOI: 10.1126/sciadv.abe4375
A balanced Oct4 interactome is crucial for maintaining pluripotency
Abstract
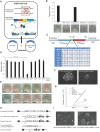
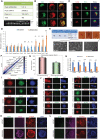
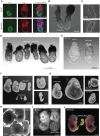
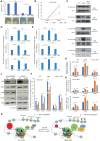
Oct4 collaborates primarily with other transcriptional factors or coregulators to maintain pluripotency. However, how Oct4 exerts its function is still unclear. Here, we show that the Oct4 linker interface mediates competing yet balanced Oct4 protein interactions that are crucial for maintaining pluripotency. Oct4 linker mutant embryonic stem cells (ESCs) show decreased expression of self-renewal genes and increased expression of differentiation genes, resulting in impaired ESC self-renewal and early embryonic development. The linker mutation interrupts the balanced Oct4 interactome. In mutant ESCs, the interaction between Oct4 and Klf5 is decreased. In contrast, interactions between Oct4 and Cbx1, Ctr9, and Cdc73 are increased, disrupting the epigenetic state of ESCs. Control of the expression level of Klf5, Cbx1, or Cdc73 rebalances the Oct4 interactome and rescues the pluripotency of linker mutant ESCs, indicating that such factors interact with Oct4 competitively. Thus, we provide previously unidentified molecular insights into how Oct4 maintains pluripotency.
Figures


References
-
- Li M., Belmonte J. C., Ground rules of the pluripotency gene regulatory network. Nat. Rev. Genet. 18, 180–191 (2017). - PubMed
-
- Rosner M. H., Vigano M. A., Ozato K., Timmons P. M., Poirier F., Rigby P. W., Staudt L. M., A POU-domain transcription factor in early stem cells and germ cells of the mammalian embryo. Nature 345, 686–692 (1990). - PubMed
-
- Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Schöler H., Smith A., Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95, 379–391 (1998). - PubMed
-
- Niwa H., Miyazaki J., Smith A. G., Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24, 372–376 (2000). - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

