Release of CHK-2 from PPM-1.D anchorage schedules meiotic entry
- PMID: 35171669
- PMCID: PMC8849337
- DOI: 10.1126/sciadv.abl8861
Release of CHK-2 from PPM-1.D anchorage schedules meiotic entry
Abstract
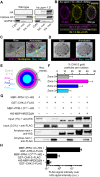
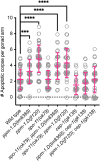
Transition from the stem/progenitor cell fate to meiosis is mediated by several redundant posttranscriptional regulatory pathways in Caenorhabditis elegans. Interfering with all three branches causes tumorous germ lines. SCFPROM-1 comprises one branch and mediates a scheduled degradation step at entry into meiosis. prom-1 mutants show defects in the timely initiation of meiotic prophase I events, resulting in high rates of embryonic lethality. Here, we identify the phosphatase PPM-1.D/Wip1 as crucial substrate for PROM-1. We report that PPM-1.D antagonizes CHK-2 kinase, a key regulator for meiotic prophase initiation, including DNA double-strand breaks, chromosome pairing, and synaptonemal complex formation. We propose that PPM-1.D controls the amount of active CHK-2 via both catalytic and noncatalytic activities; notably, noncatalytic regulation seems to be crucial at meiotic entry. PPM-1.D sequesters CHK-2 at the nuclear periphery, and programmed SCFPROM-1-mediated degradation of PPM-1.D liberates the kinase and promotes meiotic entry.
Figures




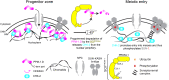
References
-
- Hansen D., Wilson-Berry L., Dang T., Schedl T., Control of the proliferation versus meiotic development decision in the C. elegans germline through regulation of GLD-1 protein accumulation. Development 131, 93–104 (2004). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

