Site-Specific Synthesis of N4-Acetylcytidine in RNA Reveals Physiological Duplex Stabilization
- PMID: 35172571
- PMCID: PMC11583671
- DOI: 10.1021/jacs.1c11985
Site-Specific Synthesis of N4-Acetylcytidine in RNA Reveals Physiological Duplex Stabilization
Abstract
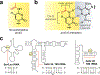
N4-Acetylcytidine (ac4C) is a post-transcriptional modification of RNA that is conserved across all domains of life. All characterized sites of ac4C in eukaryotic RNA occur in the central nucleotide of a 5'-CCG-3' consensus sequence. However, the thermodynamic consequences of cytidine acetylation in this context have never been assessed due to its challenging synthesis. Here, we report the synthesis and biophysical characterization of ac4C in its endogenous eukaryotic sequence context. First, we develop a synthetic route to homogeneous RNAs containing electrophilic acetyl groups. Next, we use thermal denaturation to interrogate the biochemical effects of ac4C on duplex stability and mismatch discrimination in a native sequence found in human rRNA. Finally, we demonstrate the ability of this chemistry to incorporate ac4C into the complex modification landscape of human tRNA and use duplex melting to highlight an enforcing role for ac4C in this unique sequence context. By enabling ex vivo biophysical analyses of nucleic acid acetylation in its physiological sequence context, these studies establish a chemical foundation for understanding the function of a universally conserved nucleobase in biology and disease.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

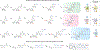


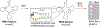
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

