Migrators within migrators: exploring transposable element dynamics in the monarch butterfly, Danaus plexippus
- PMID: 35172896
- PMCID: PMC8848866
- DOI: 10.1186/s13100-022-00263-5
Migrators within migrators: exploring transposable element dynamics in the monarch butterfly, Danaus plexippus
Abstract
Background: Lepidoptera (butterflies and moths) are an important model system in ecology and evolution. A high-quality chromosomal genome assembly is available for the monarch butterfly (Danaus plexippus), but it lacks an in-depth transposable element (TE) annotation, presenting an opportunity to explore monarch TE dynamics and the impact of TEs on shaping the monarch genome.
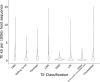
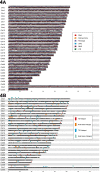
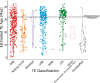
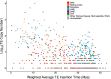
Results: We find 6.21% of the monarch genome is comprised of TEs, a reduction of 6.85% compared to the original TE annotation performed on the draft genome assembly. Monarch TE content is low compared to two closely related species with available genomes, Danaus chrysippus (33.97% TE) and Danaus melanippus (11.87% TE). The biggest TE contributions to genome size in the monarch are LINEs and Penelope-like elements, and three newly identified families, r2-hero_dPle (LINE), penelope-1_dPle (Penelope-like), and hase2-1_dPle (SINE), collectively contribute 34.92% of total TE content. We find evidence of recent TE activity, with two novel Tc1 families rapidly expanding over recent timescales (tc1-1_dPle, tc1-2_dPle). LINE fragments show signatures of genomic deletions indicating a high rate of TE turnover. We investigate associations between TEs and wing colouration and immune genes and identify a three-fold increase in TE content around immune genes compared to other host genes.
Conclusions: We provide a detailed TE annotation and analysis for the monarch genome, revealing a considerably smaller TE contribution to genome content compared to two closely related Danaus species with available genome assemblies. We identify highly successful novel DNA TE families rapidly expanding over recent timescales, and ongoing signatures of both TE expansion and removal highlight the dynamic nature of repeat content in the monarch genome. Our findings also suggest that insect immune genes are promising candidates for future interrogation of TE-mediated host adaptation.
Keywords: Butterfly; Danaus plexippus; Genome Evolution; Genomic Deletion; Lepidoptera; Repeat; TE Annotation; Transposon.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Schrader L, Schmitz J. The impact of transposable elements in adaptive evolution. Mol Ecol. 2018. 10.1111/mec.14794. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

