Proteomic analysis of archival breast cancer clinical specimens identifies biological subtypes with distinct survival outcomes
- PMID: 35173148
- PMCID: PMC8850446
- DOI: 10.1038/s41467-022-28524-0
Proteomic analysis of archival breast cancer clinical specimens identifies biological subtypes with distinct survival outcomes
Abstract
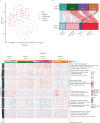
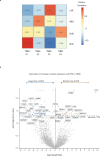
Despite advances in genomic classification of breast cancer, current clinical tests and treatment decisions are commonly based on protein level information. Formalin-fixed paraffin-embedded (FFPE) tissue specimens with extended clinical outcomes are widely available. Here, we perform comprehensive proteomic profiling of 300 FFPE breast cancer surgical specimens, 75 of each PAM50 subtype, from patients diagnosed in 2008-2013 (n = 178) and 1986-1992 (n = 122) with linked clinical outcomes. These two cohorts are analyzed separately, and we quantify 4214 proteins across all 300 samples. Within the aggressive PAM50-classified basal-like cases, proteomic profiling reveals two groups with one having characteristic immune hot expression features and highly favorable survival. Her2-Enriched cases separate into heterogeneous groups differing by extracellular matrix, lipid metabolism, and immune-response features. Within 88 triple-negative breast cancers, four proteomic clusters display features of basal-immune hot, basal-immune cold, mesenchymal, and luminal with disparate survival outcomes. Our proteomic analysis characterizes the heterogeneity of breast cancer in a clinically-applicable manner, identifies potential biomarkers and therapeutic targets, and provides a resource for clinical breast cancer classification.
© 2022. The Author(s).
Conflict of interest statement
S.K.L.C. reports receiving consulting fees from Novartis Pharma, Pfizer, Hoffman LaRoche, Merck, AstraZeneca, Eli Lilly. T.O.N. played a role in the development of the PAM50 gene expression classifier, which has been licensed to Veracyte Technologies. The other authors declare no competing interests.
Figures





References
-
- Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

