The AUTOTAC chemical biology platform for targeted protein degradation via the autophagy-lysosome system
- PMID: 35173167
- PMCID: PMC8850458
- DOI: 10.1038/s41467-022-28520-4
The AUTOTAC chemical biology platform for targeted protein degradation via the autophagy-lysosome system
Erratum in
-
Author Correction: The AUTOTAC chemical biology platform for targeted protein degradation via the autophagy-lysosome system.Nat Commun. 2022 Apr 12;13(1):2108. doi: 10.1038/s41467-022-29845-w. Nat Commun. 2022. PMID: 35413961 Free PMC article. No abstract available.
Abstract
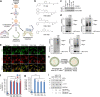
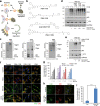
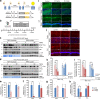
Targeted protein degradation allows targeting undruggable proteins for therapeutic applications as well as eliminating proteins of interest for research purposes. While several degraders that harness the proteasome or the lysosome have been developed, a technology that simultaneously degrades targets and accelerates cellular autophagic flux is still missing. In this study, we develop a general chemical tool and platform technology termed AUTOphagy-TArgeting Chimera (AUTOTAC), which employs bifunctional molecules composed of target-binding ligands linked to autophagy-targeting ligands. AUTOTACs bind the ZZ domain of the otherwise dormant autophagy receptor p62/Sequestosome-1/SQSTM1, which is activated into oligomeric bodies in complex with targets for their sequestration and degradation. We use AUTOTACs to degrade various oncoproteins and degradation-resistant aggregates in neurodegeneration at nanomolar DC50 values in vitro and in vivo. AUTOTAC provides a platform for selective proteolysis in basic research and drug development.
© 2022. The Author(s).
Conflict of interest statement
Seoul National University and AUTOTAC Bio, Inc. have filed patent applications (C.H.J., H.Y.K., M.J.L., A.J.H., S.G., J.E.N., H.T.K., and Y.T.K.; US 17/262,157 undergoing continuation-in-part, PCT/KR2019/009205 under examination; proof-of-concept AUTOTAC platform) based on the results of this study. The remaining authors declare no competing interests.
Figures




Comment in
-
News and views : AUTOTACs join the arena for targeted protein degradation.Arch Toxicol. 2022 Jul;96(7):2143-2144. doi: 10.1007/s00204-022-03312-3. Epub 2022 May 17. Arch Toxicol. 2022. PMID: 35577987 Free PMC article. No abstract available.
-
Targeted protein degradation via the autophagy-lysosome system: AUTOTAC (AUTOphagy-TArgeting Chimera).Autophagy. 2022 Sep;18(9):2259-2262. doi: 10.1080/15548627.2022.2091338. Epub 2022 Jun 26. Autophagy. 2022. PMID: 35722947 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

