A machine learning model for predicting deterioration of COVID-19 inpatients
- PMID: 35173197
- PMCID: PMC8850417
- DOI: 10.1038/s41598-022-05822-7
A machine learning model for predicting deterioration of COVID-19 inpatients
Abstract
The COVID-19 pandemic has been spreading worldwide since December 2019, presenting an urgent threat to global health. Due to the limited understanding of disease progression and of the risk factors for the disease, it is a clinical challenge to predict which hospitalized patients will deteriorate. Moreover, several studies suggested that taking early measures for treating patients at risk of deterioration could prevent or lessen condition worsening and the need for mechanical ventilation. We developed a predictive model for early identification of patients at risk for clinical deterioration by retrospective analysis of electronic health records of COVID-19 inpatients at the two largest medical centers in Israel. Our model employs machine learning methods and uses routine clinical features such as vital signs, lab measurements, demographics, and background disease. Deterioration was defined as a high NEWS2 score adjusted to COVID-19. In the prediction of deterioration within the next 7-30 h, the model achieved an area under the ROC curve of 0.84 and an area under the precision-recall curve of 0.74. In external validation on data from a different hospital, it achieved values of 0.76 and 0.7, respectively.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
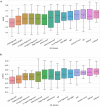

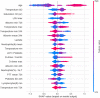

References
-
- COVID-19 Map - Johns Hopkins Coronavirus Resource Center. https://coronavirus.jhu.edu/map.html. Accessed Jun. 04, 2021.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

