Complete mitogenomes of four Trichiurus species: A taxonomic review of the T.lepturus species complex
- PMID: 35173516
- PMCID: PMC8810657
- DOI: 10.3897/zookeys.1084.71576
Complete mitogenomes of four Trichiurus species: A taxonomic review of the T.lepturus species complex
Abstract
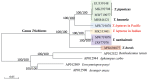
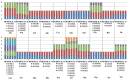
Four Trichiurus species, T.japonicus, T.lepturus, T.nanhaiensis, and T.brevis, from the coasts of the China Seas, have been identified and their entire mitochondrial genomes (mitogenomes) have been sequenced by next-generation sequencing technology. A comparative analysis of five mitogenomes was conducted, including the mitogenome of T.gangeticus. The mitogenomes contained 16.568-16.840 bp and encoded 36 typical mitochondrial genes (13 protein-coding, 2 ribosomal RNA-coding, and 21 transfer RNA-coding genes) and two typical noncoding control regions. Although tRNAPro is absent from Trichiurus mitogenomes, when compared with the 22 tRNAs reported in other vertebrates, the gene arrangements in the mitogenomes of the studied species are consistent with those in most teleost mitogenomes. The full-length sequences and protein-coding genes (PCGs) in the mitogenomes of the five species had obvious AT biases and negative GC skew values. Our study indicate that the specimens in the Indian Ocean are neither T.lepturus nor T.nanhaiensis but they are T.gangeticus; the Trichiurus species composition in the Indian Ocean is totally different from that in Pacific and Atlantic oceans; there are at least two Trichiurus species in Indian Ocean; and the worldwide systematics and diversity of the genus Trichiurus need to be reviewed.
Keywords: Characterization; Trichiurus; mitogenome; molecular tool; phylogeny; taxonomy.
Mu-Rong Yi, Kui-Ching Hsu, Sui Gu, Xiong-Bo He, Zhi-Sen Luo, Hung-Du Lin, Yun-Rong Yan.
Figures






References
-
- Ahti PA, Coleman RR, DiBattista JD, Berumen ML, Rocha LA, Bowen BW. (2016) Phylogeography of Indo-Pacific reef fishes: sister wrasses Corisgaimard and C.cuvieri in the Red Sea, Indian Ocean and Pacific Ocean. Journal of Biogeography 43: 1103–1115. 10.1111/jbi.12712 - DOI
-
- Arrondo NV, Gomes-dos-Santos A, Marcote ER, Perez M, Froufe E, Castro LFC. (2020) A new gene order in the mitochondrial genome of the deep-sea diaphanous hatchet fish Sternoptyxdiaphana Hermann, 1781 (Stomiiformes: Sternoptychidae). Mitochondrial DNA B Resource 5: 2859–2861. 10.1080/23802359.2020.1790325 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
