Investigation of the Importance of Protein 3D Structure for Assessing Conservation of Lysine Acetylation Sites in Protein Homologs
- PMID: 35173693
- PMCID: PMC8843374
- DOI: 10.3389/fmicb.2021.805181
Investigation of the Importance of Protein 3D Structure for Assessing Conservation of Lysine Acetylation Sites in Protein Homologs
Abstract
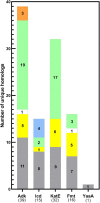
Acetylation is a protein post-translational modification (PTM) that can affect a variety of cellular processes. In bacteria, two PTM Nε-acetylation mechanisms have been identified: non-enzymatic/chemical acetylation via acetyl phosphate or acetyl coenzyme A and enzymatic acetylation via protein acetyltransferases. Prior studies have shown that extensive acetylation of Nε-lysine residues of numerous proteins from a variety of bacteria occurs via non-enzymatic acetylation. In Escherichia coli, new Nε-lysine acetyltransferases (KATs) that enzymatically acetylate other proteins have been identified, thus expanding the repertoire of protein substrates that are potentially regulated by acetylation. Therefore, we designed a study to leverage the wealth of structural data in the Protein Data Bank (PDB) to determine: (1) the 3D location of lysine residues on substrate proteins that are acetylated by E. coli KATs, and (2) investigate whether these residues are conserved on 3D structures of their homologs. Five E. coli KAT substrate proteins that were previously identified as being acetylated by YiaC and had 3D structures in the PDB were selected for further analysis: adenylate kinase (Adk), isocitrate dehydrogenase (Icd), catalase HPII (KatE), methionyl-tRNA formyltransferase (Fmt), and a peroxide stress resistance protein (YaaA). We methodically compared over 350 protein structures of these E. coli enzymes and their homologs; to accurately determine lysine residue conservation requires a strategy that incorporates both flexible structural alignments and visual inspection. Moreover, our results revealed discrepancies in conclusions about lysine residue conservation in homologs when examining linear amino acid sequences compared to 3D structures.
Keywords: Escherichia coli acetylation; Gcn5-related N-acetyltransferase (GNAT); N-epsilon lysine acetylation; conservation of protein acetylation sites; lysine acetylation; non-enzymatic acetylation; protein acetylation.
Copyright © 2022 Jew, Le, Amaral, Ta, Nguyen May, Law, Adelstein and Kuhn.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






Similar articles
-
Identification of Novel Protein Lysine Acetyltransferases in Escherichia coli.mBio. 2018 Oct 23;9(5):e01905-18. doi: 10.1128/mBio.01905-18. mBio. 2018. PMID: 30352934 Free PMC article.
-
Assays to Study Enzymatic and Non-Enzymatic Protein Lysine Acetylation In Vitro.Curr Protoc. 2021 Nov;1(11):e277. doi: 10.1002/cpz1.277. Curr Protoc. 2021. PMID: 34748287
-
Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation.PLoS One. 2014 Apr 22;9(4):e94816. doi: 10.1371/journal.pone.0094816. eCollection 2014. PLoS One. 2014. PMID: 24756028 Free PMC article.
-
Bacterial protein acetylation: mechanisms, functions, and methods for study.Front Cell Infect Microbiol. 2024 Jul 4;14:1408947. doi: 10.3389/fcimb.2024.1408947. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39027134 Free PMC article. Review.
-
Versatility of ARD1/NAA10-mediated protein lysine acetylation.Exp Mol Med. 2018 Jul 27;50(7):1-13. doi: 10.1038/s12276-018-0100-7. Exp Mol Med. 2018. PMID: 30054464 Free PMC article. Review.
Cited by
-
HDAC6 Degrades nsp8 of Porcine Deltacoronavirus through Deacetylation and Ubiquitination to Inhibit Viral Replication.J Virol. 2023 May 31;97(5):e0037523. doi: 10.1128/jvi.00375-23. Epub 2023 May 3. J Virol. 2023. PMID: 37133375 Free PMC article.
-
Acetylation, ADP-ribosylation and methylation of malate dehydrogenase.Essays Biochem. 2024 Oct 3;68(2):199-212. doi: 10.1042/EBC20230080. Essays Biochem. 2024. PMID: 38994669 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

