Gut Microbiota Composition Is Related to AD Pathology
- PMID: 35173707
- PMCID: PMC8843078
- DOI: 10.3389/fimmu.2021.794519
Gut Microbiota Composition Is Related to AD Pathology
Abstract
Introduction: Several studies have reported alterations in gut microbiota composition of Alzheimer's disease (AD) patients. However, the observed differences are not consistent across studies. We aimed to investigate associations between gut microbiota composition and AD biomarkers using machine learning models in patients with AD dementia, mild cognitive impairment (MCI) and subjective cognitive decline (SCD).
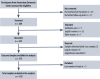
Materials and methods: We included 170 patients from the Amsterdam Dementia Cohort, comprising 33 with AD dementia (66 ± 8 years, 46%F, mini-mental state examination (MMSE) 21[19-24]), 21 with MCI (64 ± 8 years, 43%F, MMSE 27[25-29]) and 116 with SCD (62 ± 8 years, 44%F, MMSE 29[28-30]). Fecal samples were collected and gut microbiome composition was determined using 16S rRNA sequencing. Biomarkers of AD included cerebrospinal fluid (CSF) amyloid-beta 1-42 (amyloid) and phosphorylated tau (p-tau), and MRI visual scores (medial temporal atrophy, global cortical atrophy, white matter hyperintensities). Associations between gut microbiota composition and dichotomized AD biomarkers were assessed with machine learning classification models. The two models with the highest area under the curve (AUC) were selected for logistic regression, to assess associations between the 20 best predicting microbes and the outcome measures from these machine learning models while adjusting for age, sex, BMI, diabetes, medication use, and MMSE.
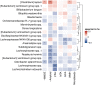
Results: The machine learning prediction for amyloid and p-tau from microbiota composition performed best with AUCs of 0.64 and 0.63. Highest ranked microbes included several short chain fatty acid (SCFA)-producing species. Higher abundance of [Clostridium] leptum and lower abundance of [Eubacterium] ventriosum group spp., Lachnospiraceae spp., Marvinbryantia spp., Monoglobus spp., [Ruminococcus] torques group spp., Roseburia hominis, and Christensenellaceae R-7 spp., was associated with higher odds of amyloid positivity. We found associations between lower abundance of Lachnospiraceae spp., Lachnoclostridium spp., Roseburia hominis and Bilophila wadsworthia and higher odds of positive p-tau status.
Conclusions: Gut microbiota composition was associated with amyloid and p-tau status. We extend on recent studies that observed associations between SCFA levels and AD CSF biomarkers by showing that lower abundances of SCFA-producing microbes were associated with higher odds of positive amyloid and p-tau status.
Keywords: Alzheimer’s disease; MRI; P-tau; amyloid beta; gut microbiota; microbiome.
Copyright © 2022 Verhaar, Hendriksen, de Leeuw, Doorduijn, van Leeuwenstijn, Teunissen, Barkhof, Scheltens, Kraaij, van Duijn, Nieuwdorp, Muller and van der Flier.
Conflict of interest statement
CT received grants from the European Commission, the Dutch Research Council (ZonMW), Association of Frontotemporal Dementia/Alzheimer’s Drug Discovery Foundation, The Weston Brain Institute, Alzheimer Netherlands. CT has a collaboration contract with ADx Neurosciences, performed contract research or received grants from Probiodrug, Biogen, Esai, Toyama, Janssen prevention center, Boehringer, AxonNeurosciences, Fujirebio, EIP farma, PeopleBio, and Roche. FB is a consultant for Biogen-Idec, Bayer-Schering, Merck-Serono, Roche, Combinostics and IXICO; has received sponsorship from European Commission–Horizon 2020, National Institute for Health Research–University College London Hospitals Biomedical Research Centre, Novartis, and Merck; and serves on the editorial boards of Radiology, Neuroradiology, Multiple Sclerosis Journal, and Neurology. PS has received consultancy/speaker fees from Lilly, GE Healthcare, Novartis, Sanofi, Nutricia, Probiodrug, Biogen, Roche, Avraham, and EIP Pharma. PS has acquired grant support from GE Healthcare, Danone Research, Piramal, and MERCK. All funding was paid to the institution. MN is part of the Scientific Advisory Board of Caelus Health, The Netherlands and Kaleido Biosciences, USA. However, none of these are directly relevant to the current paper. WF received research funding from ZonMW, NWO, EU-FP7, EU-JPND, Alzheimer Nederland, CardioVascular Onderzoek Nederland, Health~Holland, Topsector Life Sciences & Health, stichting Dioraphte, Gieskes-Strijbis fonds, stichting Equilibrio, Pasman stichting, Biogen MA Inc, Boehringer Ingelheim, Life-MI, AVID, Roche BV, Fujifilm, Combinostics. WF holds the Pasman chair. WF is recipient of ABOARD, which is a public-private partnership receiving funding from ZonMW (#73305095007) and Health~Holland, Topsector Life Sciences & Health (PPP-allowance; #LSHM20106). She has performed contract research for Biogen MA Inc, and Boehringer Ingelheim. She has been an invited speaker at Boehringer Ingelheim, Biogen MA Inc, Danone, Eisai, WebMD Neurology (Medscape). WF is consultant to Oxford Health Policy Forum CIC, Roche, and Biogen MA Inc. WF participated in an advisory board of Biogen MA Inc and Roche. WF was associate editor of Alzheimer, Research & Therapy in 2020/2021. WF is associate editor at Brain. All funding was paid to the institution. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

