Structural Basis for Unusual TCR CDR3β Usage Against an Immunodominant HIV-1 Gag Protein Peptide Restricted to an HLA-B*81:01 Molecule
- PMID: 35173732
- PMCID: PMC8841528
- DOI: 10.3389/fimmu.2022.822210
Structural Basis for Unusual TCR CDR3β Usage Against an Immunodominant HIV-1 Gag Protein Peptide Restricted to an HLA-B*81:01 Molecule
Abstract
In HIV infection, some closely associated human leukocyte antigen (HLA) alleles are correlated with distinct clinical outcomes although presenting the same HIV epitopes. The mechanism that underpins this observation is still unknown, but may be due to the essential features of HLA alleles or T cell receptors (TCR). In this study, we investigate how T18A TCR, which is beneficial for a long-term control of HIV in clinic, recognizes immunodominant Gag epitope TL9 (TPQDLTML180-188) from HIV in the context of the antigen presenting molecule HLA-B*81:01. We found that T18A TCR exhibits differential recognition for TL9 restricted by HLA-B*81:01. Furthermore, via structural and biophysical approaches, we observed that TL9 complexes with HLA-B*81:01 undergoes no conformational change after TCR engagement. Remarkably, the CDR3β in T18A complexes does not contact with TL9 at all but with intensive contacts to HLA-B*81:01. The binding kinetic data of T18A TCR revealed that this TCR can recognize TL9 epitope and several mutant versions, which might explain the correlation of T18A TCR with better clinic outcomes despite the relative high mutation rate of HIV. Collectively, we provided a portrait of how CD8+ T cells engage in HIV-mediated T cell response.
Keywords: CD8+ T cells; HIV; HLA; T cell receptor; antigen presentation.
Copyright © 2022 Liu, Lei, San, Yang, Paek, Xia, Chen and Yin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
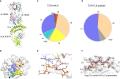
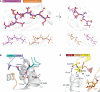

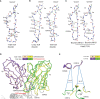

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

