Versatile role of miR-24/24-1*/24-2* expression in cancer and other human diseases
- PMID: 35173828
- PMCID: PMC8829624
Versatile role of miR-24/24-1*/24-2* expression in cancer and other human diseases
Abstract
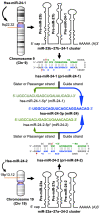
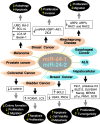
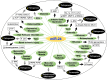
MiRNAs (miRs) have been proven to be well-validated therapeutic targets. Emerging evidence has demonstrated that intricate, intrinsic and paradoxical functions of miRs are context-dependent because of their multiple upstream regulators, broad spectrum of downstream molecular targets and distinct expression in various tissues, organs and disease states. Targeted therapy has become an emerging field of research. One key for the development of successful miR-based/targeted therapy is to acquire integrated knowledge of its regulatory network and its association with disease phenotypes to identify critical nodes of the underlying pathogenesis. Herein, we systematically summarized the comprehensive role of miR-24-3p (miR-24), along with its passenger strands miR-24-1-5p* (miR-24-1) and miR-24-2-5p* (miR-24-2), emphasizing their microenvironment, intracellular targets, and associated gene networks and regulatory phenotypes in 18 different cancer types and 13 types of other disorders. MiR-24 targets and regulates numerous genes in various cancer types and enhances the expression of several oncogenes (e.g., cMyc, BCL2 and HIF1), which are challenging in terms of druggability. In contrast, several tumor suppressor proteins (p21 and p53) have been reported to be downregulated by miR-24. MiR-24 also regulates the cell cycle and is associated with numerous cancer hallmarks such as apoptosis, proliferation, metastasis, invasion, angiogenesis, autophagy, drug resistance and other diseases pathogenesis. Overall, miR-24 plays an emerging role in the diagnosis, prognosis and pathobiology of various diseases. MiR-24 is a potential target for targeted therapy in the era of precision medicine, which expands the landscape of targetable macromolecules, including undruggable proteins.
Keywords: MiR-24/24-1*/24-2*; regulatory role in cancer and other diseases; target genes and regulatory networks; therapeutic target.
AJTR Copyright © 2022.
Conflict of interest statement
None.
Figures


Similar articles
-
Dual strands of the miR-223 duplex (miR-223-5p and miR-223-3p) inhibit cancer cell aggressiveness: targeted genes are involved in bladder cancer pathogenesis.J Hum Genet. 2018 May;63(5):657-668. doi: 10.1038/s10038-018-0437-8. Epub 2018 Mar 14. J Hum Genet. 2018. PMID: 29540855
-
RNA-sequence-based microRNA expression signature in breast cancer: tumor-suppressive miR-101-5p regulates molecular pathogenesis.Mol Oncol. 2020 Feb;14(2):426-446. doi: 10.1002/1878-0261.12602. Epub 2019 Dec 29. Mol Oncol. 2020. PMID: 31755218 Free PMC article.
-
Molecular pathogenesis of breast cancer: impact of miR-99a-5p and miR-99a-3p regulation on oncogenic genes.J Hum Genet. 2021 May;66(5):519-534. doi: 10.1038/s10038-020-00865-y. Epub 2020 Nov 12. J Hum Genet. 2021. PMID: 33177704
-
Network pharmacology-based identification of miRNA expression of Astragalus membranaceus in the treatment of diabetic nephropathy.Medicine (Baltimore). 2022 Feb 4;101(5):e28747. doi: 10.1097/MD.0000000000028747. Medicine (Baltimore). 2022. PMID: 35119030 Free PMC article.
-
Dysregulated expression and functions of microRNA-330 in cancers: A potential therapeutic target.Biomed Pharmacother. 2022 Feb;146:112600. doi: 10.1016/j.biopha.2021.112600. Epub 2021 Dec 27. Biomed Pharmacother. 2022. PMID: 34968919
Cited by
-
Suppression of RBFox2 by Multiple MiRNAs in Pressure Overload-Induced Heart Failure.Int J Mol Sci. 2023 Jan 9;24(2):1283. doi: 10.3390/ijms24021283. Int J Mol Sci. 2023. PMID: 36674797 Free PMC article.
-
Keratinocyte-derived extracellular vesicles in painful diabetic neuropathy.Neurobiol Pain. 2024 Dec 17;17:100176. doi: 10.1016/j.ynpai.2024.100176. eCollection 2025 Jan-Jun. Neurobiol Pain. 2024. PMID: 39811188 Free PMC article.
-
Biomarkers for Oral Squamous Cell Carcinoma (miR-24, miR-200, and miR-34): Screening and Detection MicroRNA.Asian Pac J Cancer Prev. 2024 Jul 1;25(7):2265-2269. doi: 10.31557/APJCP.2024.25.7.2265. Asian Pac J Cancer Prev. 2024. PMID: 39068557 Free PMC article.
-
The interplay of p16INK4a and non-coding RNAs: bridging cellular senescence, aging, and cancer.Biogerontology. 2025 Feb 5;26(2):50. doi: 10.1007/s10522-025-10194-2. Biogerontology. 2025. PMID: 39907830 Review.
-
Discovering common pathogenetic processes between periodontitis and Alzheimer's disease by bioinformatics and system biology approach.BMC Oral Health. 2024 Sep 12;24(1):1074. doi: 10.1186/s12903-024-04775-9. BMC Oral Health. 2024. PMID: 39266981 Free PMC article.
References
-
- Lee RC, Feinbaun RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. - PubMed
-
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the microprocessor complex. Nature. 2004;432:231–235. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
