The Prognostic and Predictive Role of Xeroderma Pigmentosum Gene Expression in Melanoma
- PMID: 35174087
- PMCID: PMC8841870
- DOI: 10.3389/fonc.2022.810058
The Prognostic and Predictive Role of Xeroderma Pigmentosum Gene Expression in Melanoma
Abstract
Background: Assessment of immune-specific markers is a well-established approach for predicting the response to immune checkpoint inhibitors (ICIs). Promising candidates as ICI predictive biomarkers are the DNA damage response pathway genes. One of those pathways, which are mainly responsible for the repair of DNA damage caused by ultraviolet radiation, is the nucleotide excision repair (NER) pathway. Xeroderma pigmentosum (XP) is a hereditary disease caused by mutations of eight different genes of the NER pathway, or POLH, here together named the nine XP genes. Anecdotal evidence indicated that XP patients with melanoma or other skin tumors responded impressively well to anti-PD-1 ICIs. Hence, we analyzed the expression of the nine XP genes as prognostic and anti-PD-1 ICI predictive biomarkers in melanoma.
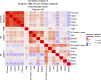
Methods: We assessed mRNA gene expression in the TCGA-SKCM dataset (n = 445) and two pooled clinical melanoma cohorts of anti-PD-1 ICI (n = 75). In TCGA-SKCM, we applied hierarchical clustering on XP genes to reveal clusters, further utilized as XP cluster scores. In addition, out of 18 predefined genes representative of a T cell inflamed tumor microenvironment, the TIS score was calculated. Besides these scores, the XP genes, immune-specific single genes (CD8A, CXCL9, CD274, and CXCL13) and tumor mutational burden (TMB) were cross-correlated. Survival analysis in TCGA-SKCM was conducted for the selected parameters. Lastly, the XP response prediction value was calculated for the two pooled anti-PD-1 cohorts by classification models.
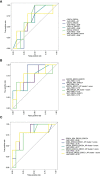
Results: In TCGA-SKCM, expression of the XP genes was divided into two clusters, inversely correlated with immune-specific markers. A higher ERCC3 expression was associated with improved survival, particularly in younger patients. The constructed models utilizing XP genes, and the XP cluster scores outperformed the immune-specific gene-based models in predicting response to anti-PD-1 ICI in the pooled clinical cohorts. However, the best prediction was achieved by combining the immune-specific gene CD274 with three XP genes from both clusters.
Conclusion: Our results suggest pre-therapeutic XP gene expression as a potential marker to improve the prediction of anti-PD-1 response in melanoma.
Keywords: DNA damage response; RNA-seq; anti-PD-1; biomarker; gene expression; melanoma; nucleotide excision repair; xeroderma pigmentosum.
Copyright © 2022 Fischer, Hamed, Emmert, Wolkenhauer, Fuellen and Thiem.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources
Research Materials

