Fiber-Like Organization as a Basic Principle for Euchromatin Higher-Order Structure
- PMID: 35174159
- PMCID: PMC8841976
- DOI: 10.3389/fcell.2021.784440
Fiber-Like Organization as a Basic Principle for Euchromatin Higher-Order Structure
Abstract
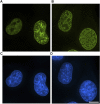
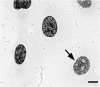
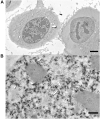
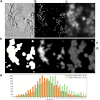
A detailed understanding of the principles of the structural organization of genetic material is of great importance for elucidating the mechanisms of differential regulation of genes in development. Modern ideas about the spatial organization of the genome are based on a microscopic analysis of chromatin structure and molecular data on DNA-DNA contact analysis using Chromatin conformation capture (3C) technology, ranging from the "polymer melt" model to a hierarchical folding concept. Heterogeneity of chromatin structure depending on its functional state and cell cycle progression brings another layer of complexity to the interpretation of structural data and requires selective labeling of various transcriptional states under nondestructive conditions. Here, we use a modified approach for replication timing-based metabolic labeling of transcriptionally active chromatin for ultrastructural analysis. The method allows pre-embedding labeling of optimally structurally preserved chromatin, thus making it compatible with various 3D-TEM techniques including electron tomography. By using variable pulse duration, we demonstrate that euchromatic genomic regions adopt a fiber-like higher-order structure of about 200 nm in diameter (chromonema), thus providing support for a hierarchical folding model of chromatin organization as well as the idea of transcription and replication occurring on a highly structured chromatin template.
Keywords: electron tomography; euchromatin; higher-order chromatin folding; replication; transcription.
Copyright © 2022 Zakirov, Sosnovskaya, Ryumina, Kharybina, Strelkova, Zhironkina, Golyshev, Moiseenko and Kireev.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
LinkOut - more resources
Full Text Sources

