A Ferroptosis-Related Gene Prognostic Index to Predict Temozolomide Sensitivity and Immune Checkpoint Inhibitor Response for Glioma
- PMID: 35174170
- PMCID: PMC8842730
- DOI: 10.3389/fcell.2021.812422
A Ferroptosis-Related Gene Prognostic Index to Predict Temozolomide Sensitivity and Immune Checkpoint Inhibitor Response for Glioma
Abstract
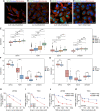
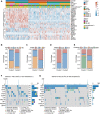
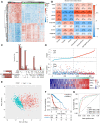
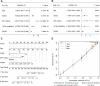
Background: Gliomas are highly lethal brain tumors. Despite multimodality therapy with surgery, radiotherapy, chemotherapy, and immunotherapy, glioma prognosis remains poor. Ferroptosis is a crucial tumor suppressor mechanism that has been proven to be effective in anticancer therapy. However, the implications of ferroptosis on the clinical prognosis, chemotherapy, and immune checkpoint inhibitor (ICI) therapy for patients with glioma still need elucidation. Methods: Consensus clustering revealed two distinct ferroptosis-related subtypes based on the Cancer Genome Atlas (TCGA) glioma dataset (n = 663). Subsequently, the ferroptosis-related gene prognostic index (FRGPI) was constructed by weighted gene co-expression network analysis (WGCNA) and "stepAIC" algorithms and validated with the Chinese Glioma Genome Atlas (CGGA) dataset (n = 404). Subsequently, the correlation among clinical, molecular, and immune features and FRGPI was analyzed. Next, the temozolomide sensitivity and ICI response for glioma were predicted using the "pRRophetic" and "TIDE" algorithms, respectively. Finally, candidate small molecular drugs were defined using the connectivity map database based on FRGPI. Results: The FRGPI was established based on the HMOX1, TFRC, JUN, and SOCS1 genes. The distribution of FRGPI varied significantly among the different ferroptosis-related subtypes. Patients with high FRGPI had a worse overall prognosis than patients with low FRGPI, consistent with the results in the CGGA dataset. The final results showed that high FRGPI was characterized by more aggressive phenotypes, high PD-L1 expression, high tumor mutational burden score, and enhanced temozolomide sensitivity; low FRGPI was associated with less aggressive phenotypes, high microsatellite instability score, and stronger response to immune checkpoint blockade. In addition, the infiltration of memory resting CD4+ T cells, regulatory T cells, M1 macrophages, M2 macrophages, and neutrophils was positively correlated with FRGPI. In contrast, plasma B cells and naïve CD4+ T cells were negatively correlated. A total of 15 potential small molecule compounds (such as depactin, physostigmine, and phenacetin) were identified. Conclusion: FRGPI is a promising gene panel for predicting the prognosis, immune characteristics, temozolomide sensitivity, and ICI response in patients with glioma.
Keywords: ferroptosis; ferroptosis-based anticancer therapy; glioma; immune checkpoint inhibitor; immunotherapy; temozolomide; tumor microenvironment.
Copyright © 2022 Cai, Liang, Zhan, Zeng, Lin, Xu, Xue, Xu, Chai, Mao, Song, Han, Xiao, Song and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
A novel ferroptosis-related gene prognostic index for prognosis and response to immunotherapy in patients with prostate cancer.Front Endocrinol (Lausanne). 2022 Aug 10;13:975623. doi: 10.3389/fendo.2022.975623. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 36034466 Free PMC article.
-
One Ferroptosis-Related Gene-Pair Signature Serves as an Original Prognostic Biomarker in Lung Adenocarcinoma.Front Genet. 2022 Mar 16;13:841712. doi: 10.3389/fgene.2022.841712. eCollection 2022. Front Genet. 2022. PMID: 35368652 Free PMC article.
-
A Novel lncRNA Panel Related to Ferroptosis, Tumor Progression, and Microenvironment is a Robust Prognostic Indicator for Glioma Patients.Front Cell Dev Biol. 2021 Dec 7;9:788451. doi: 10.3389/fcell.2021.788451. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34950662 Free PMC article.
-
Ferroptosis-related NFE2L2 and NOX4 Genes are Potential Risk Prognostic Biomarkers and Correlated with Immunogenic Features in Glioma.Cell Biochem Biophys. 2023 Mar;81(1):7-17. doi: 10.1007/s12013-022-01124-x. Epub 2023 Jan 11. Cell Biochem Biophys. 2023. PMID: 36627482 Free PMC article. Review.
-
Differential genes and scoring criteria among immunogenomic clusters of lower-grade gliomas.Int Immunopharmacol. 2021 Dec;101(Pt B):108376. doi: 10.1016/j.intimp.2021.108376. Epub 2021 Nov 20. Int Immunopharmacol. 2021. PMID: 34815191 Review.
Cited by
-
CYP2E1 overexpression protects COS-7 cancer cells against ferroptosis.Res Sq [Preprint]. 2023 Mar 22:rs.3.rs-2702878. doi: 10.21203/rs.3.rs-2702878/v1. Res Sq. 2023. PMID: 36993697 Free PMC article. Preprint.
-
N-glycosylation of ACTRIIB enhances protein stability leading to rapid cell proliferation and strong resistance to docetaxel in nasopharyngeal carcinoma.Braz J Med Biol Res. 2025 Feb 3;58:e14368. doi: 10.1590/1414-431X2024e14368. eCollection 2025. Braz J Med Biol Res. 2025. PMID: 39907411 Free PMC article.
-
Ferroptosis in glioma therapy: advancements in sensitizing strategies and the complex tumor-promoting roles.Brain Res. 2024 Oct 1;1840:149045. doi: 10.1016/j.brainres.2024.149045. Epub 2024 May 29. Brain Res. 2024. PMID: 38821335 Review.
-
Ferroptosis Modulation: Potential Therapeutic Target for Glioblastoma Treatment.Int J Mol Sci. 2022 Jun 21;23(13):6879. doi: 10.3390/ijms23136879. Int J Mol Sci. 2022. PMID: 35805884 Free PMC article. Review.
-
Multifaceted bioinformatic analysis of m6A-related ferroptosis and its link with gene signatures and tumour-infiltrating immune cells in gliomas.J Cell Mol Med. 2024 Sep;28(17):e70060. doi: 10.1111/jcmm.70060. J Cell Mol Med. 2024. PMID: 39248438 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

