Intra- and extra-cranial BCOR-ITD tumours are separate entities within the BCOR-rearranged family
- PMID: 35174661
- PMCID: PMC8977275
- DOI: 10.1002/cjp2.255
Intra- and extra-cranial BCOR-ITD tumours are separate entities within the BCOR-rearranged family
Abstract
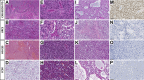
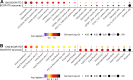
BCOR-ITD tumours form an emerging family of aggressive entities with an internal tandem duplication (ITD) in the last exon of the BCOR gene. The family includes cerebral tumours, termed central nervous system BCOR-ITD (CNS BCOR-ITD), and sarcomatous types described in the kidney as clear cell sarcoma of the kidney (CCSK), in the endometrium as high-grade endometrial stromal sarcoma, and in the bone and soft tissue as undifferentiated round cell sarcoma or primitive myxoid mesenchymal tumour of infancy. Based on a series of 33 retrospective cases, including 10 CNS BCOR-ITD and 23 BCOR-ITD sarcomas, we interrogated the homogeneity of the entity regarding clinical, radiological, and histopathological findings, and molecular signatures. Whole-transcriptomic sequencing and DNA methylation profiling were used for unsupervised clustering. BCOR-ITD tumours mostly affected young children with a median age at diagnosis of 2.1 years (range 0-62.4). Median overall survival was 3.9 years and progression-free survival was 1.4 years. This dismal prognosis is shared among tumours in all locations except CCSK. Histopathological review revealed marked differences between CNS BCOR-ITD and BCOR-ITD sarcomas. These two groups were consistently segregated by unsupervised clustering of expression (n = 22) and DNA methylation (n = 21) data. Proximity between the two groups may result from common somatic changes within key pathways directly related to the novel activity of the ITD itself. Conversely, comparison of gene signatures with single-cell RNA-Seq atlases suggests that the distinction between BCOR-ITD sarcomas and CNS BCOR-ITD may result from differences in cells of origin.
Keywords: BCOR-ITD sarcomas; CCSK; CNS BCOR-ITD; ESS; clustering; methylome; transcriptome.
© 2022 The Authors. The Journal of Pathology: Clinical Research published by The Pathological Society of Great Britain and Ireland & John Wiley & Sons, Ltd.
Figures



References
-
- Ueno‐Yokohata H, Okita H, Nakasato K, et al. Consistent in‐frame internal tandem duplications of BCOR characterize clear cell sarcoma of the kidney. Nat Genet 2015; 47: 861–863. - PubMed
-
- Karlsson J, Valind A, Gisselsson D. BCOR internal tandem duplication and YWHAE‐NUTM2B/E fusion are mutually exclusive events in clear cell sarcoma of the kidney. Genes Chromosomes Cancer 2016; 55: 120–123. - PubMed
-
- Kenny C, Bausenwein S, Lazaro A, et al. Mutually exclusive BCOR internal tandem duplications and YWHAE‐NUTM2 fusions in clear cell sarcoma of kidney: not the full story. J Pathol 2016; 238: 617–620. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

