Neutral Loss Mass Spectral Data Enhances Molecular Similarity Analysis in METLIN
- PMID: 35174708
- PMCID: PMC10131246
- DOI: 10.1021/jasms.1c00343
Neutral Loss Mass Spectral Data Enhances Molecular Similarity Analysis in METLIN
Abstract
Neutral loss (NL) spectral data presents a mirror of MS2 data and is a valuable yet largely untapped resource for molecular discovery and similarity analysis. Tandem mass spectrometry (MS2) data is effective for the identification of known molecules and the putative identification of novel, previously uncharacterized molecules (unknowns). Yet, MS2 data alone is limited in characterizing structurally related molecules. To facilitate unknown identification and complement the METLIN-MS2 fragment ion database for characterizing structurally related molecules, we have created a MS2 to NL converter as a part of the METLIN platform. The converter has been used to transform METLIN's MS2 data into a neutral loss database (METLIN-NL) on over 860 000 individual molecular standards. The platform includes both the MS2 to NL converter and a graphical user interface enabling comparative analyses between MS2 and NL data. Examples of NL spectral data are shown with oxylipin analogues and two structurally related statin molecules to demonstrate NL spectra and their ability to help characterize structural similarity. Mirroring MS2 data to generate NL spectral data offers a unique dimension for chemical and metabolite structure characterization.
Figures

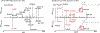
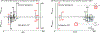
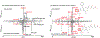
References
-
- Guijas C; Montenegro-Burke JR; Domingo-Almenara X; Palermo A; Warth B; Hermann G; Koellensperger G; Huan T; Uritboonthai W; Aisporna AE; Wolan DW; Spilker ME; Benton HP; Siuzdak G METLIN: A technology platform for identifying knowns and unknowns. Anal Chem 2018, 90(5), 3156–64. 10.1021/acs.analchem.7b04424 - DOI - PMC - PubMed

