Visualizing the next frontiers in wine yeast research
- PMID: 35175339
- PMCID: PMC8916113
- DOI: 10.1093/femsyr/foac010
Visualizing the next frontiers in wine yeast research
Abstract
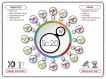
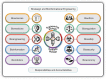
A range of game-changing biodigital and biodesign technologies are coming of age all around us, transforming our world in complex ways that are hard to predict. Not a day goes by without news of how data-centric engineering, algorithm-driven modelling, and biocyber technologies-including the convergence of artificial intelligence, machine learning, automated robotics, quantum computing, and genome editing-will change our world. If we are to be better at expecting the unexpected in the world of wine, we need to gain deeper insights into the potential and limitations of these technological developments and advances along with their promise and perils. This article anticipates how these fast-expanding bioinformational and biodesign toolkits might lead to the creation of synthetic organisms and model systems, and ultimately new understandings of biological complexities could be achieved. A total of four future frontiers in wine yeast research are discussed in this article: the construction of fully synthetic yeast genomes, including minimal genomes; supernumerary pan-genome neochromosomes; synthetic metagenomes; and synthetic yeast communities. These four concepts are at varying stages of development with plenty of technological pitfalls to overcome before such model chromosomes, genomes, strains, and yeast communities could illuminate some of the ill-understood aspects of yeast resilience, fermentation performance, flavour biosynthesis, and ecological interactions in vineyard and winery settings. From a winemaker's perspective, some of these ideas might be considered as far-fetched and, as such, tempting to ignore. However, synthetic biologists know that by exploring these futuristic concepts in the laboratory could well forge new research frontiers to deepen our understanding of the complexities of consistently producing fine wines with different fermentation processes from distinctive viticultural terroirs. As the saying goes in the disruptive technology industry, it take years to create an overnight success. The purpose of this article is neither to glorify any of these concepts as a panacea to all ills nor to crucify them as a danger to winemaking traditions. Rather, this article suggests that these proposed research endeavours deserve due consideration because they are likely to cast new light on the genetic blind spots of wine yeasts, and how they interact as communities in vineyards and wineries. Future-focussed research is, of course, designed to be subject to revision as new data and technologies become available. Successful dislodging of old paradigms with transformative innovations will require open-mindedness and pragmatism, not dogmatism-and this can make for a catch-22 situation in an archetypal traditional industry, such as the wine industry, with its rich territorial and socio-cultural connotations.
Keywords: Saccharomyces cerevisiae; minimal genome; pan-genome; supernumerary neochromosome; synthetic communities; synthetic genome; wine yeast.
© The Author(s) 2022. Published by Oxford University Press on behalf of FEMS.
Figures





References
-
- Almeida P, Barbosa R, Zalar Pet al. A population genomics insight into Mediterranean origins of wine yeast domestication. Mol Ecol. 2015;24:5412–27. - PubMed
-
- Banerjee S, Schlaeppi K, van der Heijden MGA.. Keystone taxa as drivers of microbiome structure and functioning. Nat Rev Microbiol. 2018;16:567–76. - PubMed
-
- Belda I., Ruiz J, Santos Aet al. Saccharomyces cerevisiae . Trends Genet. 2019;35:956–7. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

