Systematic profiling of antigen bias in humoral response against SARS-CoV-2
- PMID: 35176329
- PMCID: PMC8842411
- DOI: 10.1016/j.virusres.2022.198711
Systematic profiling of antigen bias in humoral response against SARS-CoV-2
Abstract
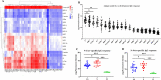
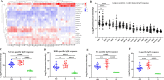
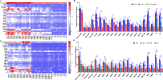
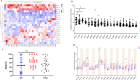
We know little about the antigen bias in SARS-CoV-2 humoral response and the epitopes of spike recognized by the immune system in asymptomatic (AS) patients and symptomatic (S) patients. Here, we used a microarray to evaluate the humoral immune response in the sera collected from 33 COVID-19-recovered patients up to 1 year. We found that the levels of IgG and IgM induced by the 23 proteins differed significantly in the same patients, and were able to distinguish AS and S patients. The N- and S-specific antibodies were detected even at 12 months after onset. Five epitopes were identified to be associated with the clinical adverse events, and three peptides located in RBD. Overall, this study presents a systemic view of the SARS-CoV-2 specific IgG and IgM responses between AS and S recovered patients and provide insights to promote precise development of SARS-CoV-2 vaccines.
Keywords: Antibodies; Antigen bias; Epitopes; Proteome microarray; SARS-CoV-2.
Copyright © 2022 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
Evaluation of Spike Protein Epitopes by Assessing the Dynamics of Humoral Immune Responses in Moderate COVID-19.Front Immunol. 2022 Mar 18;13:770982. doi: 10.3389/fimmu.2022.770982. eCollection 2022. Front Immunol. 2022. PMID: 35371042 Free PMC article.
-
Longitudinal immune profiling reveals dominant epitopes mediating long-term humoral immunity in COVID-19-convalescent individuals.J Allergy Clin Immunol. 2022 Apr;149(4):1225-1241. doi: 10.1016/j.jaci.2022.01.005. Epub 2022 Jan 21. J Allergy Clin Immunol. 2022. PMID: 35074422 Free PMC article.
-
Characterization of SARS-CoV-2-Specific Humoral and Cellular Immune Responses Induced by Inactivated COVID-19 Vaccines in a Real-World Setting.Front Immunol. 2021 Dec 22;12:802858. doi: 10.3389/fimmu.2021.802858. eCollection 2021. Front Immunol. 2021. PMID: 35003131 Free PMC article.
-
Neutralization of SARS-CoV-2 requires antibodies against conformational receptor-binding domain epitopes.Allergy. 2022 Jan;77(1):230-242. doi: 10.1111/all.15066. Epub 2021 Sep 22. Allergy. 2022. PMID: 34453317 Free PMC article.
-
SARS-CoV-2 recombinant proteins stimulate distinct cellular and humoral immune response profiles in samples from COVID-19 convalescent patients.Clinics (Sao Paulo). 2021 Dec 6;76:e3548. doi: 10.6061/clinics/2021/e3548. eCollection 2021. Clinics (Sao Paulo). 2021. PMID: 34878034 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

