Molecular basis of force-pCa relation in MYL2 cardiomyopathy mice: Role of the super-relaxed state of myosin
- PMID: 35177471
- PMCID: PMC8872785
- DOI: 10.1073/pnas.2110328119
Molecular basis of force-pCa relation in MYL2 cardiomyopathy mice: Role of the super-relaxed state of myosin
Abstract
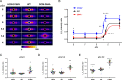
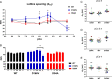
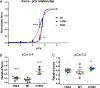
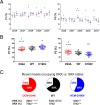
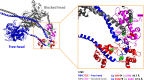
In this study, we investigated the role of the super-relaxed (SRX) state of myosin in the structure-function relationship of sarcomeres in the hearts of mouse models of cardiomyopathy-bearing mutations in the human ventricular regulatory light chain (RLC, MYL2 gene). Skinned papillary muscles from hypertrophic (HCM-D166V) and dilated (DCM-D94A) cardiomyopathy models were subjected to small-angle X-ray diffraction simultaneously with isometric force measurements to obtain the interfilament lattice spacing and equatorial intensity ratios (I11/I10) together with the force-pCa relationship over a full range of [Ca2+] and at a sarcomere length of 2.1 μm. In parallel, we studied the effect of mutations on the ATP-dependent myosin energetic states. Compared with wild-type (WT) and DCM-D94A mice, HCM-D166V significantly increased the Ca2+ sensitivity of force and left shifted the I11/I10-pCa relationship, indicating an apparent movement of HCM-D166V cross-bridges closer to actin-containing thin filaments, thereby allowing for their premature Ca2+ activation. The HCM-D166V model also disrupted the SRX state and promoted an SRX-to-DRX (super-relaxed to disordered relaxed) transition that correlated with an HCM-linked phenotype of hypercontractility. While this dysregulation of SRX ↔ DRX equilibrium was consistent with repositioning of myosin motors closer to the thin filaments and with increased force-pCa dependence for HCM-D166V, the DCM-D94A model favored the energy-conserving SRX state, but the structure/function-pCa data were similar to WT. Our results suggest that the mutation-induced redistribution of myosin energetic states is one of the key mechanisms contributing to the development of complex clinical phenotypes associated with human HCM-D166V and DCM-D94A mutations.
Keywords: equatorial intensity ratio; interfilament lattice spacing; isometric force; super-relaxed state of myosin; transgenic RLC mice.
Conflict of interest statement
The authors declare no competing interest.
Figures
References
-
- Richard P, et al. Hypertrophic cardiomyopathy: Distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation 107, 2227–2232 (2003), and erratum in Circulation 109, 3258 (2004). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

