Affinity maturation for an optimal balance between long-term immune coverage and short-term resource constraints
- PMID: 35177475
- PMCID: PMC8872716
- DOI: 10.1073/pnas.2113512119
Affinity maturation for an optimal balance between long-term immune coverage and short-term resource constraints
Abstract
In order to target threatening pathogens, the adaptive immune system performs a continuous reorganization of its lymphocyte repertoire. Following an immune challenge, the B cell repertoire can evolve cells of increased specificity for the encountered strain. This process of affinity maturation generates a memory pool whose diversity and size remain difficult to predict. We assume that the immune system follows a strategy that maximizes the long-term immune coverage and minimizes the short-term metabolic costs associated with affinity maturation. This strategy is defined as an optimal decision process on a finite dimensional phenotypic space, where a preexisting population of cells is sequentially challenged with a neutrally evolving strain. We show that the low specificity and high diversity of memory B cells-a key experimental result-can be explained as a strategy to protect against pathogens that evolve fast enough to escape highly potent but narrow memory. This plasticity of the repertoire drives the emergence of distinct regimes for the size and diversity of the memory pool, depending on the density of de novo responding cells and on the mutation rate of the strain. The model predicts power-law distributions of clonotype sizes observed in data and rationalizes antigenic imprinting as a strategy to minimize metabolic costs while keeping good immune protection against future strains.
Keywords: B cell repertoire; affinity maturation; optimal decision theory; population dynamics.
Conflict of interest statement
The authors declare no competing interest.
Figures
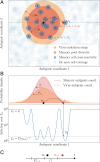
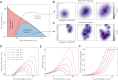
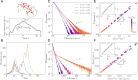

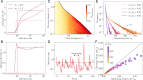
References
-
- Nieuwenhuis P., Opstelten D., Functional anatomy of germinal centers. Am. J. Anat. 170, 421–435 (1984). - PubMed
-
- Victora G. D., Nussenzweig M. C., Germinal centers. Annu. Rev. Immunol. 30, 429–457 (2012). - PubMed
-
- Eisen H. N., Siskind G. W., Variations in affinities of antibodies during the immune response. Biochemistry 3, 996–1008 (1964). - PubMed
-
- Weisel F., Shlomchik M., Memory B cells of mice and humans. Annu. Rev. Immunol. 35, 255–284 (2017). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

