RBBP6 activates the pre-mRNA 3' end processing machinery in humans
- PMID: 35177536
- PMCID: PMC8887125
- DOI: 10.1101/gad.349223.121
RBBP6 activates the pre-mRNA 3' end processing machinery in humans
Abstract
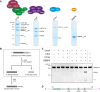
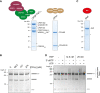
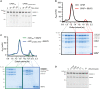
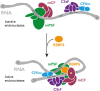
3' end processing of most human mRNAs is carried out by the cleavage and polyadenylation specificity factor (CPSF; CPF in yeast). Endonucleolytic cleavage of the nascent pre-mRNA defines the 3' end of the mature transcript, which is important for mRNA localization, translation, and stability. Cleavage must therefore be tightly regulated. Here, we reconstituted specific and efficient 3' endonuclease activity of human CPSF with purified proteins. This required the seven-subunit CPSF as well as three additional protein factors: cleavage stimulatory factor (CStF), cleavage factor IIm (CFIIm), and, importantly, the multidomain protein RBBP6. Unlike its yeast homolog Mpe1, which is a stable subunit of CPF, RBBP6 does not copurify with CPSF and is recruited in an RNA-dependent manner. Sequence and mutational analyses suggest that RBBP6 interacts with the WDR33 and CPSF73 subunits of CPSF. Thus, it is likely that the role of RBBP6 is conserved from yeast to humans. Overall, our data are consistent with CPSF endonuclease activation and site-specific pre-mRNA cleavage being highly controlled to maintain fidelity in mRNA processing.
Keywords: RNA; endonuclease; gene expression; polyadenylation.
© 2022 Boreikaite et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
