Reconstitution of 3' end processing of mammalian pre-mRNA reveals a central role of RBBP6
- PMID: 35177537
- PMCID: PMC8887130
- DOI: 10.1101/gad.349217.121
Reconstitution of 3' end processing of mammalian pre-mRNA reveals a central role of RBBP6
Abstract
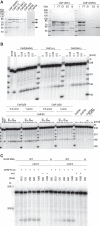
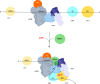
The 3' ends of almost all eukaryotic mRNAs are generated in an essential two-step processing reaction: endonucleolytic cleavage of an extended precursor followed by the addition of a poly(A) tail. By reconstituting the reaction from overproduced and purified proteins, we provide a minimal list of 14 polypeptides that are essential and two that are stimulatory for RNA processing. In a reaction depending on the polyadenylation signal AAUAAA, the reconstituted system cleaves pre-mRNA at a single preferred site corresponding to the one used in vivo. Among the proteins, cleavage factor I stimulates cleavage but is not essential, consistent with its prominent role in alternative polyadenylation. RBBP6 is required, with structural data showing it to contact and presumably activate the endonuclease CPSF73 through its DWNN domain. The C-terminal domain of RNA polymerase II is dispensable. ATP, but not its hydrolysis, supports RNA cleavage by binding to the hClp1 subunit of cleavage factor II with submicromolar affinity.
Keywords: 3′ processing; CPSF; RBBP6; RNA cleavage; RNA processing; poly(A) polymerase; polyadenylation.
© 2022 Schmidt et al.; Published by Cold Spring Harbor Laboratory Press.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
