ASPP2 maintains the integrity of mechanically stressed pseudostratified epithelia during morphogenesis
- PMID: 35177595
- PMCID: PMC8854694
- DOI: 10.1038/s41467-022-28590-4
ASPP2 maintains the integrity of mechanically stressed pseudostratified epithelia during morphogenesis
Abstract
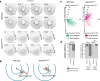
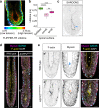
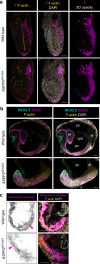
During development, pseudostratified epithelia undergo large scale morphogenetic events associated with increased mechanical stress. Using a variety of genetic and imaging approaches, we uncover that in the mouse E6.5 epiblast, where apical tension is highest, ASPP2 safeguards tissue integrity. It achieves this by preventing the most apical daughter cells from delaminating apically following division events. In this context, ASPP2 maintains the integrity and organisation of the filamentous actin cytoskeleton at apical junctions. ASPP2 is also essential during gastrulation in the primitive streak, in somites and in the head fold region, suggesting that it is required across a wide range of pseudostratified epithelia during morphogenetic events that are accompanied by intense tissue remodelling. Finally, our study also suggests that the interaction between ASPP2 and PP1 is essential to the tumour suppressor function of ASPP2, which may be particularly relevant in the context of tissues that are subject to increased mechanical stress.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Norden C. Pseudostratified epithelia — cell biology, diversity and roles in organ formation at a glance. J. Cell Sci. 2017;130:1859–1863. - PubMed
-
- Hoijman, E., Rubbini, D., Colombelli, J. & Alsina, B. Mitotic cell rounding and epithelial thinning regulate lumen growth and shape. Nat. Commun. 6, 7355 (2015). - PubMed
-
- Kondo T, Hayashi S. Mitotic cell rounding accelerates epithelial invagination. Nature. 2013;494:125–129. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

