Detecting SARS-CoV-2 lineages and mutational load in municipal wastewater and a use-case in the metropolitan area of Thessaloniki, Greece
- PMID: 35177697
- PMCID: PMC8854625
- DOI: 10.1038/s41598-022-06625-6
Detecting SARS-CoV-2 lineages and mutational load in municipal wastewater and a use-case in the metropolitan area of Thessaloniki, Greece
Abstract
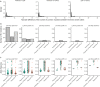
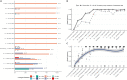
The COVID-19 pandemic represents an unprecedented global crisis necessitating novel approaches for, amongst others, early detection of emerging variants relating to the evolution and spread of the virus. Recently, the detection of SARS-CoV-2 RNA in wastewater has emerged as a useful tool to monitor the prevalence of the virus in the community. Here, we propose a novel methodology, called lineagespot, for the monitoring of mutations and the detection of SARS-CoV-2 lineages in wastewater samples using next-generation sequencing (NGS). Our proposed method was tested and evaluated using NGS data produced by the sequencing of 14 wastewater samples from the municipality of Thessaloniki, Greece, covering a 6-month period. The results showed the presence of SARS-CoV-2 variants in wastewater data. lineagespot was able to record the evolution and rapid domination of the Alpha variant (B.1.1.7) in the community, and allowed the correlation between the mutations evident through our approach and the mutations observed in patients from the same area and time periods. lineagespot is an open-source tool, implemented in R, and is freely available on GitHub and registered on bio.tools.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Jahn, K. et al. Detection of SARS-CoV-2 Variants in Switzerland by Genomic Analysis of Wastewater Samples. 10.1101/2021.01.08.21249379 (2021).
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

