YAP/TAZ drives cell proliferation and tumour growth via a polyamine-eIF5A hypusination-LSD1 axis
- PMID: 35177822
- PMCID: PMC8930503
- DOI: 10.1038/s41556-022-00848-5
YAP/TAZ drives cell proliferation and tumour growth via a polyamine-eIF5A hypusination-LSD1 axis
Abstract
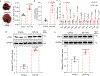
Metabolic reprogramming is central to oncogene-induced tumorigenesis by providing the necessary building blocks and energy sources, but how oncogenic signalling controls metabolites that play regulatory roles in driving cell proliferation and tumour growth is less understood. Here we show that oncogene YAP/TAZ promotes polyamine biosynthesis by activating the transcription of the rate-limiting enzyme ornithine decarboxylase 1. The increased polyamine levels, in turn, promote the hypusination of eukaryotic translation factor 5A (eIF5A) to support efficient translation of histone demethylase LSD1, a transcriptional repressor that mediates a bulk of YAP/TAZ-downregulated genes including tumour suppressors in YAP/TAZ-activated cells. Accentuating the importance of the YAP/TAZ-polyamine-eIF5A hypusination-LSD1 axis, inhibiting polyamine biosynthesis or LSD1 suppressed YAP/TAZ-induced cell proliferation and tumour growth. Given the frequent upregulation of YAP/TAZ activity and polyamine levels in diverse cancers, our identification of YAP/TAZ as an upstream regulator and LSD1 as a downstream effector of the oncometabolite polyamine offers a molecular framework in which oncogene-induced metabolic and epigenetic reprogramming coordinately drives tumorigenesis, and suggests potential therapeutic strategies in YAP/TAZ- or polyamine-dependent human malignancies.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
The authors have no financial and non-financial competing interests.
Figures






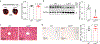
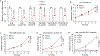


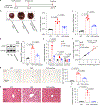
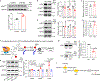



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

