Metabolomic and microbiome profiling reveals personalized risk factors for coronary artery disease
- PMID: 35177859
- PMCID: PMC12365913
- DOI: 10.1038/s41591-022-01686-6
Metabolomic and microbiome profiling reveals personalized risk factors for coronary artery disease
Abstract
Complex diseases, such as coronary artery disease (CAD), are often multifactorial, caused by multiple underlying pathological mechanisms. Here, to study the multifactorial nature of CAD, we performed comprehensive clinical and multi-omic profiling, including serum metabolomics and gut microbiome data, for 199 patients with acute coronary syndrome (ACS) recruited from two major Israeli hospitals, and validated these results in a geographically distinct cohort. ACS patients had distinct serum metabolome and gut microbial signatures as compared with control individuals, and were depleted in a previously unknown bacterial species of the Clostridiaceae family. This bacterial species was associated with levels of multiple circulating metabolites in control individuals, several of which have previously been linked to an increased risk of CAD. Metabolic deviations in ACS patients were found to be person specific with respect to their potential genetic or environmental origin, and to correlate with clinical parameters and cardiovascular outcomes. Moreover, metabolic aberrations in ACS patients linked to microbiome and diet were also observed to a lesser extent in control individuals with metabolic impairment, suggesting the involvement of these aberrations in earlier dysmetabolic phases preceding clinically overt CAD. Finally, a metabolomics-based model of body mass index (BMI) trained on the non-ACS cohort predicted higher-than-actual BMI when applied to ACS patients, and the excess BMI predictions independently correlated with both diabetes mellitus (DM) and CAD severity, as defined by the number of vessels involved. These results highlight the utility of the serum metabolome in understanding the basis of risk-factor heterogeneity in CAD.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures


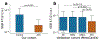



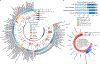



Comment in
-
Before the heart attack.Nat Med. 2022 Feb;28(2):237-238. doi: 10.1038/s41591-022-01685-7. Nat Med. 2022. PMID: 35177861 No abstract available.
-
Changes to the gut microbiota drive the progression of cardiometabolic disease.Nat Rev Cardiol. 2022 May;19(5):283. doi: 10.1038/s41569-022-00686-w. Nat Rev Cardiol. 2022. PMID: 35260795 No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

