Microbiome and metabolome features of the cardiometabolic disease spectrum
- PMID: 35177860
- PMCID: PMC8863577
- DOI: 10.1038/s41591-022-01688-4
Microbiome and metabolome features of the cardiometabolic disease spectrum
Abstract
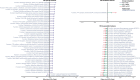
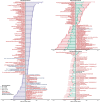
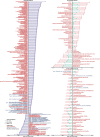
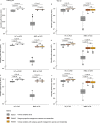
Previous microbiome and metabolome analyses exploring non-communicable diseases have paid scant attention to major confounders of study outcomes, such as common, pre-morbid and co-morbid conditions, or polypharmacy. Here, in the context of ischemic heart disease (IHD), we used a study design that recapitulates disease initiation, escalation and response to treatment over time, mirroring a longitudinal study that would otherwise be difficult to perform given the protracted nature of IHD pathogenesis. We recruited 1,241 middle-aged Europeans, including healthy individuals, individuals with dysmetabolic morbidities (obesity and type 2 diabetes) but lacking overt IHD diagnosis and individuals with IHD at three distinct clinical stages-acute coronary syndrome, chronic IHD and IHD with heart failure-and characterized their phenome, gut metagenome and serum and urine metabolome. We found that about 75% of microbiome and metabolome features that distinguish individuals with IHD from healthy individuals after adjustment for effects of medication and lifestyle are present in individuals exhibiting dysmetabolism, suggesting that major alterations of the gut microbiome and metabolome might begin long before clinical onset of IHD. We further categorized microbiome and metabolome signatures related to prodromal dysmetabolism, specific to IHD in general or to each of its three subtypes or related to escalation or de-escalation of IHD. Discriminant analysis based on specific IHD microbiome and metabolome features could better differentiate individuals with IHD from healthy individuals or metabolically matched individuals as compared to the conventional risk markers, pointing to a pathophysiological relevance of these features.
© 2022. The Author(s).
Conflict of interest statement
F.B. is a shareholder in Implexion Pharma. K.C. is a consultant for Danone Research, LNC Therapeutics and CONFO Therapeutics for work that is unassociated with the present study. K.C. has held a collaborative research contract with Danone Research in the context of the MetaCardis project. The other authors declare no competing interests.
Figures



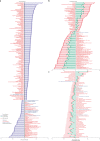
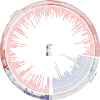
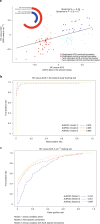


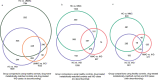



Comment in
-
Before the heart attack.Nat Med. 2022 Feb;28(2):237-238. doi: 10.1038/s41591-022-01685-7. Nat Med. 2022. PMID: 35177861 No abstract available.
References
-
- Heymsfield SB, Wadden TA. Mechanisms, pathophysiology, and management of obesity. N. Engl. J. Med. 2017;376:254–266. - PubMed
-
- Rothschild D, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–215. - PubMed
-
- Cotillard A, et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–588. - PubMed
-
- Karlsson FH, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. - PubMed
-
- Le Chatelier E, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

